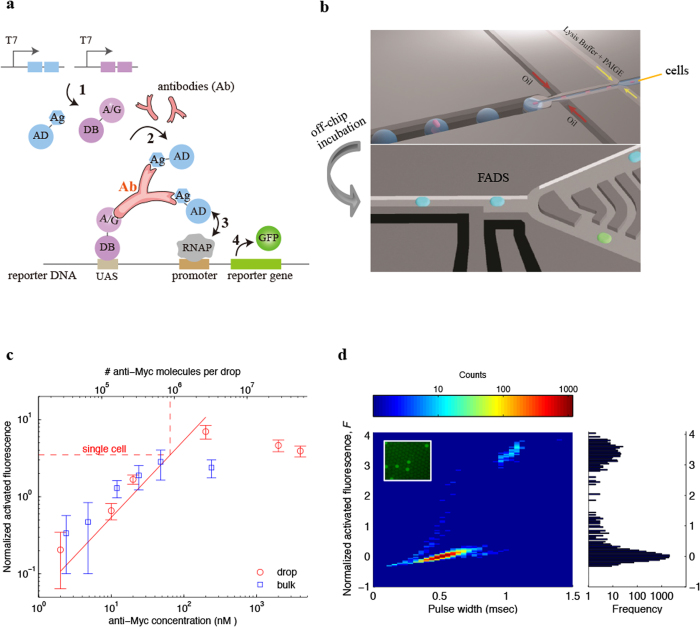
Figure 1. Detection and quantification of antibody in single hybridoma cells using drop-based PAIGE.
(a) Scheme of PAIGE for detecting an antigen (Ag)-specific antibody (Ab). Two fusion proteins, AD-Ag and A/G-DB, are constitutively expressed under T7 promoter (T7) from input DNA templates by T7 RNA polymerase (step 1). The antibody binds to both Ag and A/G, forming a ternary complex that binds to the upstream-activation sequence (UAS) on the reporter DNA and recruits AD near the promoter-bound RNA polymerase (RNAP) (step 2). AD activates RNAP (step 3) to express the reporter gene, which produces GFP (step 4). (b) The workflow of drop-based PAIGE. Single cells (megenta) are encapsulated with PAIGE and a lysis buffer in drops (blue). After off-chip incubation, drops are re-injected into a fluorescence-activated drop-sorting (FADS) device to measure their fluorescence and sort them based on a fluorescence threshold. (c) Titration curves of pure anti-myc in microwell (blue square) and drop-based (red circle) PAIGE. The dashed line shows the average number of anti-Myc molecules in a single cell. The fluorescence of anti-Myc in microwells or drops is normalized by that of microwells or drops without anti-Myc. The normalized activated fluorescence, F, is obtained by subtracting one. (d) Heat map showing the distribution of drops in terms of their normalized activated fluorescence, F, and width recorded as the PMT’s pulse duration (left). The corresponding fluorescence histogram is shown on the right. The inset fluorescence microscopy image shows that after incubation a small fraction of the drops is bright.