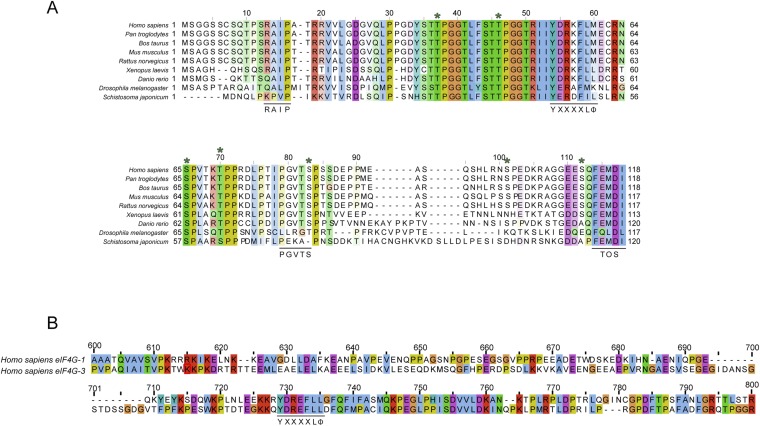
Fig. S1.
Amino acid alignments. Residues are colored according to ClustalX color scheme (57). (A) Sequence alignment of 4E-BP1. Relevant 4E-BP1 motifs are underlined (RAIP, TOS motif, YXXXXLΦ, PGVTS/T). Asterisks indicate phosphorylation sites (S35, T40, T46, T50, S65, T70, S83, S101, and S112). (B) Sequence alignment of eIF4G-1 and eIF4G-3 residues 600 and 800 containing the consensus eIF4E binding motif YXXXXLΦ (underlined).