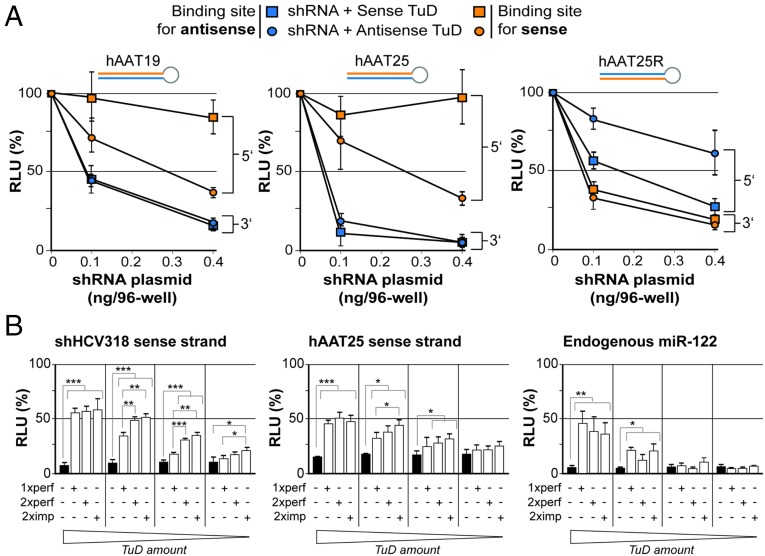
Fig. 5.
Determination of optimal shRNA and TuD design parameters. (A) Results from transfection of HEK293T cells with increasing doses of three shRNAs that differed in length and strand order but shared the same target. The cells were cotransfected with constant doses of specific TuDs and dual luciferase reporters as indicated. For each combination of TuD and reporter, luciferase reads were normalized to those measured in the absence of shRNA (set to 100%). (B) Functional analysis of three different TuD designs with one perfect, two perfect, or two imperfect binding site(s). (Left and Center) Results from HEK293T cells that were cotransfected with the indicated shRNAs and TuDs, together with a dual luciferase reporter to detect shRNA sense strand activity. (Right) Results from Huh7 cells that were cotransfected with a TuD against the 5′ arm of endogenous miR-122 and a corresponding dual luciferase reporter. In each experiment, four different TuD doses were evaluated: 100, 25, 6.25, and 1.56 ng per well (from left to right, as indicated below the graphs). Also shown in all panels are results obtained with an empty control (black bars), i.e., a TuD plasmid lacking specific binding sites for any of the three small RNAs. ***P < 0.001; **P < 0.01; *P < 0.05; two-sided t-test. Error bars in A and B represent SDs of at least three independent experiments.