Significance
The α9α10 nicotinic AChR (nAChR) subtype is a recently identified target for the development of breast cancer chemotherapeutics and analgesics, particularly to treat neuropathic pain. Structure/function analyses of antagonists of this subtype are therefore essential for the development of specific therapeutic compounds. The Conus genus is a rich source of pharmacologically active peptides, and we report here that the αO-conotoxin GeXIVA is a potent and selective antagonist of the α9α10 nAChR subtype. GeXIVA displays unique structural properties among other Conus peptides and represents a previously unidentified template for molecules active against neuropathic pain.
Keywords: αO-conotoxin GeXIVA, α9α10 nAChR, nicotinic, NMR, pain
Abstract
We identified a previously unidentified conotoxin gene from Conus generalis whose precursor signal sequence has high similarity to the O1-gene conotoxin superfamily. The predicted mature peptide, αO-conotoxin GeXIVA (GeXIVA), has four Cys residues, and its three disulfide isomers were synthesized. Previously pharmacologically characterized O1-superfamily peptides, exemplified by the US Food and Drug Administration-approved pain medication, ziconotide, contain six Cys residues and are calcium, sodium, or potassium channel antagonists. However, GeXIVA did not inhibit calcium channels but antagonized nicotinic AChRs (nAChRs), most potently on the α9α10 nAChR subtype (IC50 = 4.6 nM). Toxin blockade was voltage-dependent, and kinetic analysis of toxin dissociation indicated that the binding site of GeXIVA does not overlap with the binding site of the competitive antagonist α-conotoxin RgIA. Surprisingly, the most active disulfide isomer of GeXIVA is the bead isomer, comprising, according to NMR analysis, two well-resolved but uncoupled disulfide-restrained loops. The ribbon isomer is almost as potent but has a more rigid structure built around a short 310-helix. In contrast to most α-conotoxins, the globular isomer is the least potent and has a flexible, multiconformational nature. GeXIVA reduced mechanical hyperalgesia in the rat chronic constriction injury model of neuropathic pain but had no effect on motor performance, warranting its further investigation as a possible therapeutic agent.
Cone snails of the Conus genus are predatory marine organisms that hunt a variety of prey types, ranging from fish to worms. Venoms from these carnivorous snails are a particularly rich source of diverse peptide ligands. Due to intense evolutionary pressure, the venoms of the ∼700 known species of Conus snails represent a vast combinatorial library of neuroactive ligands. Such ligands have been developed as effective pharmacological agents for distinguishing among subtypes of nervous system receptors and ion channels (1). One example of a Conus-based peptide developed as a drug is ω-conotoxin MVIIA or ziconotide, which has US Food and Drug Administration approval as an analgesic for intractable pain. Historically, novel native peptides have been obtained by venom purification (2), but recent advances in molecular biology have provided important new ways to access and classify natural polypeptide libraries via nucleic acid analysis (3).
Nicotinic AChRs (nAChRs) are widely used throughout the animal kingdom to enable and facilitate motor movement and sensory processing, and they represent a fundamental target of predatory organisms, including cone snails. The nAChRs are ligand-gated ion channels formed from the assembly of five homologous subunits, with neuronal nAChRs assembled from a combination of α- and β-subunits. In vertebrates, there are at least 12 such subunits (α2–α10 and β2–β4) (4); some invertebrates have twice this number (5). Different combinations of nAChR subunits form subtypes of nAChRs that are used for diverse yet discrete functions. Our understanding of these functions would be greatly facilitated by the discovery and characterization of selective probes.
The α9α10 subtype of nAChR is well known for its function in mediating synaptic transmission between efferent olivocochlear fibers and hair cells of the cochlea (6). More recently, α9α10 nAChRs have been found in adrenal chromaffin cells, where they may modulate responses to stress (7). In addition, transcripts for the α9 and/or α10 subunit have been reported in a variety of nonneuronal cells, including immune cells. Reduced function of α9α10 nAChR is associated with a beneficial alteration of immune function in animal models of chronic pain and experimental autoimmune encephalomyelitis (8, 9). In vertebrates, the α9 and/or α10 subunit might be a primordial ancestor of the nAChR family (10, 11).
Conotoxins are grouped into superfamilies based on conserved endoplasmic reticulum (ER) signal peptide sequences. The current study describes the discovery and characterization of a previously unidentified O1-superfamily conotoxin, GeXIVA, representing a previously unidentified structural family of conotoxins suitable for probing the structure and function of the α9α10 nAChR.
Results
Discovery of a Clone Encoding GeXIVA from Conus generalis.
Conotoxins (i.e., disulfide-rich conopeptides) are classified according to three criteria: similarities in the ER signal sequences of the conotoxin precursors (gene superfamilies), the Cys patterns of conotoxin mature peptide regions (Cys frameworks), and specificities for pharmacological targets (pharmacological families). In an effort to discover novel conotoxins, we examined the worm-hunting cone snail Conus generalis. Specimens were collected from the South China Sea, and dissected venom ducts were used for cDNA cloning of novel conotoxins by RT-PCR.
The O1-superfamily of conotoxins is well-known for its activity on voltage-gated calcium channels (VGCCs). For example, ω-conotoxins are now widely used as defining ligands for VGCCs. There have, however, been reports of ω-conotoxins that not only potently block calcium channels but also have comparatively weak activity on nAChRs (12). We therefore wished to assess whether some O1-superfamily peptides might antagonize nAChRs. We used O1-superfamily primers to identify a cDNA clone encoding a precursor with high homology to known O1-superfamily precursors (13) (Table S1). The gene encodes a signal sequence of 22 amino acids, a proregion of 24 residues, and a C-terminal mature toxin region of 28 amino acids, some of which are predicted to be posttranslationally processed (Table S1). The mature conotoxin is separated from the proregion by the proteolytic site RLPK. Its precursor (GeXIVAP) sequence has been deposited in GenBank with accession no. KP793740.
Table S1.
Protein precursor sequences of αO-related Conus gene superfamilies
| Superfamily | Peptide | Cys framework | Precursor sequence (ER signal peptide, N-terminal proregions, ↓mature peptide, and ↓C-terminal cleavage site) | Source |
| O1 | αO-GeXIVA | C-C-C-C | MKLTCVLIITVLFLTACQLTTAVTYSRGEHKHRALMSTGTNYRLPK↓TCRSSGRYCRSPYDRRRRYCRRITDACV* | This study |
| MVIIA | C-C-CC-C-C | MKLTCVVIVAVLLLTACQLITADDSRGTQKHRALRSTTKLSTSTR↓CKGKGAKCSRLMYDCCTGSCRSGKC↓G | (86) | |
| O2 | BeB54 | C-C-CC-C-C | MEKLTILLLVAAVLMSTQALIQSDGEKRQQAKINFLS.RK↓STAESWWEGECKGWSVYCSWDWECCSGECTRYYCELW | (87) |
| O3 | CaFr179 | C-C-CC-C-C | MSGLGIMVLTLLLLVFMEASHQDAGEKQATQRDAINVRRRRSLARR↓TVTEECEEDCEDEEKHCCNTNNGPSCARLCF↓G | (87) |
| J | PIXIVA | C-C-C-C | MPSVRSVTCCCLLWMMFSVQLVTPGSPGTAQLSGHRTAR↓FPRPRICNLACRAGIGHKYPFCHCR↓GKRDAVSSSMAV | (76) |
| L | LtXIVA | C-C-C-C | MKLSVMFIVFLMLTMPMTCAGISRSATNGGEADVRAHDKAANLMALLQER↓MCPPLCKPSCTNC↓G | (78) |
| A | α-BuIA | CC-C-C | MFTVFLLVVLTTTVVSFPSDRASDGRNAAANDKASDVVTLVLK↓GCCSTPPCAVLYC↓GRRR | (88) |
| αA-PIVA | CC-C-C-C-C | MFTVFLLVVLATTVVSFTSDRASDDRNTNDKASRLLSHVVR↓GCCGSYPNAACHPCSCKDRPSYCGQ↓GR | (89) | |
| B1 | Conantokin-Br | No Cys | MQLYTYLYLLVPLVTFHLILGTGTLGHGGALTERRSTDATALK | (72) |
| PEPVLQKSAARSTDDNGKDRLTQMKRILKKRGKNAR↓GDEEYSKFIEREREAGRLDLSKFP | ||||
| B2 | Mr8.2 | C-C-C-C-C-C-C-C-C-C | MLRLITAAVLVSACLAYPQKKRTPPQTRPTSR↓ALVSQCRPCPTCRECKCRECKCRECQCRIHSCLSAWDSRGIWMRT | (73) |
| B3 | αB-VxXXIVA | C-CC-C | METLTLLWRASSSCLLVVLSHSLLRLLG↓VRCLEKSGAQPNKLFRPPCCQKGPSFARHSRCVYYTQSRE | (71) |
| C | αC-PrXA | C-C | MQTAYWVMVMMMVMWITAPLSEGGKPKLIIRGLVPNDLTPQRILRSLISGR↓TYGIYDAKPPFSCAGLRGGCVLPPNLRPKFKE↓GR | (83) |
| D | αD-VxXXA | C-CC-C-CC-C-C-C-C | MPKLEMMLLVLLIFPLSYFIAAGGQVVQVDRRGDGLAGYLQRGDR↓DVQDCQVSTPGSKWGRCCLNRVCGPMCCPASHCYCVYHRGRGHGCSC | (90) |
| S | αS-RVIIIA | C-C-C-C-C-C-C-C-C-C | MMSKMGAMFVLLLLFTLASSQQEGDVQARKTHPKREFQRILLRSGR↓KCNFDKCKGTGVYNCGESCSCEGLHSCRCTYNIGSMKSGCACICTYY | (86) |
Sequences and domain definitions are provided in ConoServer (91).
DNA sequence of GeXIVA precursor with GenBank accession no. KP793740 is as follows: ATGAAACTGACGTGCGTGTTGATCATCACCGTGCTGTTCCTGACGGCCTGTCAACTCACTACAGCTGTGACTTACTCCAGAGGTGAGCATAAGCATCGTGCTCTGATGTCAACTGGCACAAACTACAGGTTGCCCAAGACGTGCCGTAGTTCCGGTCGTTATTGTCGCTCACCTTATGATCGCCGCAGAAGATATTGCAGACGCATTACGGATGCGTGCGTATAG.
A notable feature of this previously unidentified conotoxin is that the 4-Cys framework of the predicted mature peptide differs from the 4-Cys framework of previously characterized O1-gene superfamily conotoxins, which have a 6-Cys pattern with three disulfide bridges (Table S1). Despite the high sequence identity (>60%) between their ER signal peptide sequences, previously characterized O1-superfamily peptides display different mature peptide sequences. The previously unidentified 28-residue peptide reported here is highly divergent from previously characterized conopeptides and has nine Arg residues. It was named αO-conotoxin GeXIVA (αO-GeXIVA, GeXIVA) and constitutes a previously uncharacterized structural member of the conotoxin family.
Chemical Synthesis and Folding of GeXIVA.
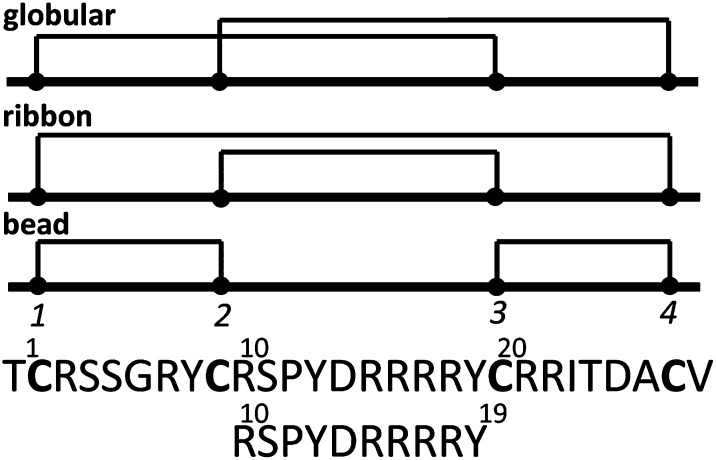
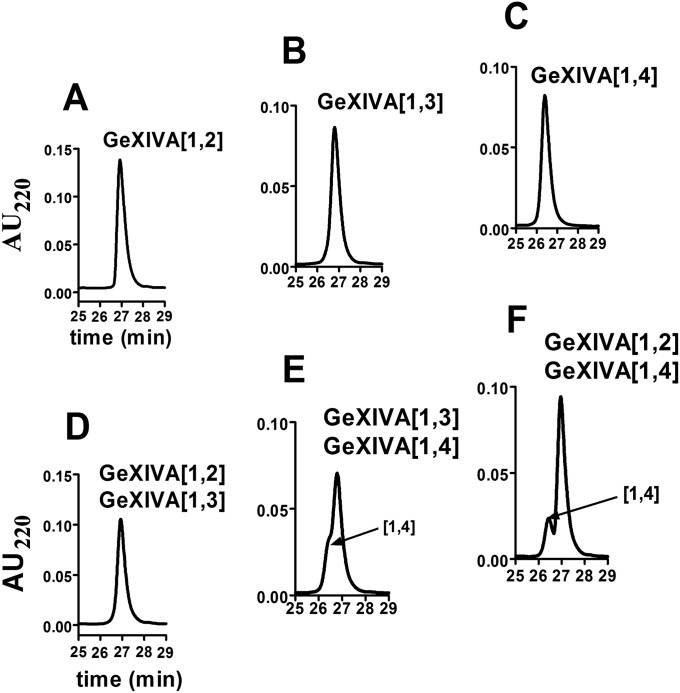
Because the disulfide connectivity of GeXIVA was not known, we synthesized homologs with the three possible disulfide bond arrangements: Cys2–Cys20, Cys9–Cys27 (globular, GeXIVA[1,3], where [1,3] indicates the connection of the first disulfide); Cys2–Cys27, Cys9–Cys20 (ribbon, GeXIVA[1,4]); and Cys2–Cys9, Cys20–Cys27 (bead, GeXIVA[1,2]) (Fig. 1). The Cys side chains were protected in pairs with orthogonal protecting groups that were removed selectively under different conditions, allowing the formation of one disulfide bridge at a time. The third and fourth, second and fourth, or second and third Cys residues were protected with acid-labile groups, which were simultaneously removed during cleavage from the resin. Ferricyanide was used to close the first disulfide bridge. Reverse-phase HPLC was used to purify the monocyclic peptide; subsequently, the acid-stable acetamidomethyl groups were removed from the remaining two Cys by iodine oxidation, which also closed the second disulfide bridge. HPLC was then used to purify the three fully folded peptide isomers (Fig. S1 A–C). Isomers were coeluted in pairs (∼1:1 ratio) to compare elution profiles (Fig. S1 D–F). The chromatograms show that GeXIVA[1,2] and GeXIVA[1,3] had the same retention time and overlap in elution profiles. MALDI-TOF MS was used to confirm the structure of the products. The observed monoisotopic mass (in daltons) of each isomer was as follows: 3,451.3 Da (GeXIVA[1,2]), 3,451.7 Da (GeXIVA[1,3]), and 3,451.9 Da (GeXIVA[1,4]), which are in good agreement with the theoretical value (3,451.96 Da). Synthetic peptides with these disulfide bond arrangements were used in all subsequent studies. Two peptides corresponding to residues 9–20 and 10–19 were also synthesized to probe the 3D structure in this region of the molecule further.
Fig. 1.
Peptide sequence of αO-conotoxin GeXIVA using a one-letter code showing the disulfide connectivities of the globular, ribbon, and bead isomers in schematic form and the sequence of the fragment peptide from R10–Y19. The globular, ribbon, and bead forms are denoted throughout the main text as GeXIVA[1,3], GeXIVA[1,4], and GeXIVA[1,2], respectively, denoting the disulfide connectivity of the first disulfide bond in each case, with the second disulfide bond defined by default. Labeling of the Cys numbering is shown for the bead isomer.
Fig. S1.
Chromatographic properties of isomers of αO-conotoxin GeXIVA. HPLC chromatograms of ∼1 nmol of GeXIVA[1,2] (A); ∼1 nmol of GeXIVA[1,3] (B); ∼1 nmol of GeXIVA[1,4] (C); coelution of ∼0.33 nmol each of GeXIVA[1,2] + GeXIVA[1,3] (D); coelution of ∼0.33 nmol each of GeXIVA[1,3] + GeXIVA[1,4] (E); and coelution of ∼0.33 nmol each of GeXIVA[1,2] + GeXIVA[1,4] (F). Note that GeXIVA[1,2] + GeXIVA[1,3] coelute, necessitating synthetic preparation by directed disulfide folding. Peptides were analyzed on RP analytical Vydac C18 HPLC using a linear gradient of 10–50% (vol/vol) buffer B60 for 40 min. Buffer B60 was 60% (vol/vol) acetonitrile, 40% (vol/vol) H20, and 0.092% trifluoroacetic acid; buffer A was 0.1% trifluoroacetic acid in H2O. A at 220 nm was monitored.
Effect of GeXIVA on nAChR ACh-Evoked Currents.
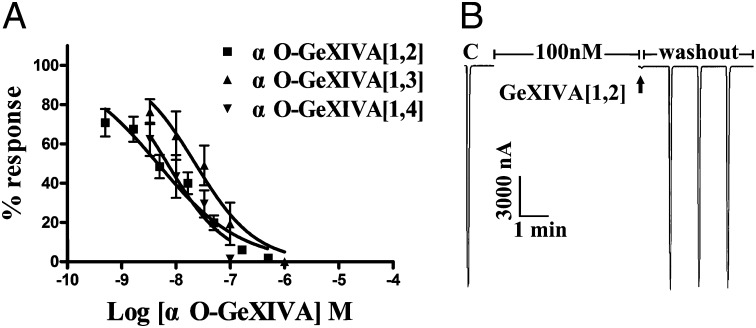
The nAChR antagonists are major components of cone snail venoms. We therefore tested the GeXIVA isomers for inhibition of rat nAChR subtypes. Pairwise combinations of rat nAChR subunits were heterologously expressed in Xenopus oocytes. Screening was initially performed at a concentration of 10 μM. The most potent activity was observed at the α9α10 nAChR subtype. Concentration response experiments on rat α9α10 nAChR were then conducted. Among the three isomers, GeXIVA[1,2] (bead form) was the most potent at the α9α10 nAChR with an IC50 of 4.6 nM (3.2–6.7 nM), followed by GeXIVA[1,4] (ribbon form) with an IC50 of 7 nM (3.6–13.4 nM). Surprisingly, GeXIVA[1,3] (the globular form) was the least active, with an IC50 of 22.7 nM (11.8–43.5 nM) (Fig. 2A). Representative responses to ACh of α9α10 nAChRs in the presence and absence of GeXIVA[1,2] are shown in Fig. 2B. Potent blockade of ACh-evoked currents was obtained with 100 nM GeXIVA[1,2] on α9α10 nAChR, and this block was rapidly reversible.
Fig. 2.
αO-conotoxin GeXIVA blocks rat α9α10 nAChRs. (A) Concentration response of GeXIVA isomers on α9α10 nAChRs. Oocytes expressing α9α10 nAChR were voltage-clamped at −70 mV and subjected to a 1-s pulse of 10 μM ACh every minute as described in Materials and Methods. The IC50s were as follows: GeXIVA[1,2], 4.6 nM (3.18–6.65 nM); GeXIVA[1,3], 22.7 nM (11.8–43.5 nM); and GeXIVA[1,4], 7 nM (3.6–13.4 nM). Data points are mean ± SEM. The numbers inside the parentheses in the legend indicate 95% confidence interval of the IC50 or Hill slope. Hill slopes were GeXIVA[1,2], 0.56 (0.44–0.69); GeXIVA[1,3], 0.78 (0.29–1.26); and GeXIVA[1,4], 0.79 (0.23–1.36). Values were from six to 12 separate oocytes. (B) Representative block of ACh response of α9α10 nAChRs by 100 nM GeXIVA[1,2]. The “C” responses are control, following which the oocyte was exposed to peptide for 5 min. The arrow denotes the first ACh response in the presence of toxin. Peptide was then washed out, and subsequent responses to ACh were measured at 1-min intervals. Blockade of α9α10 nAChRs was rapidly reversed.
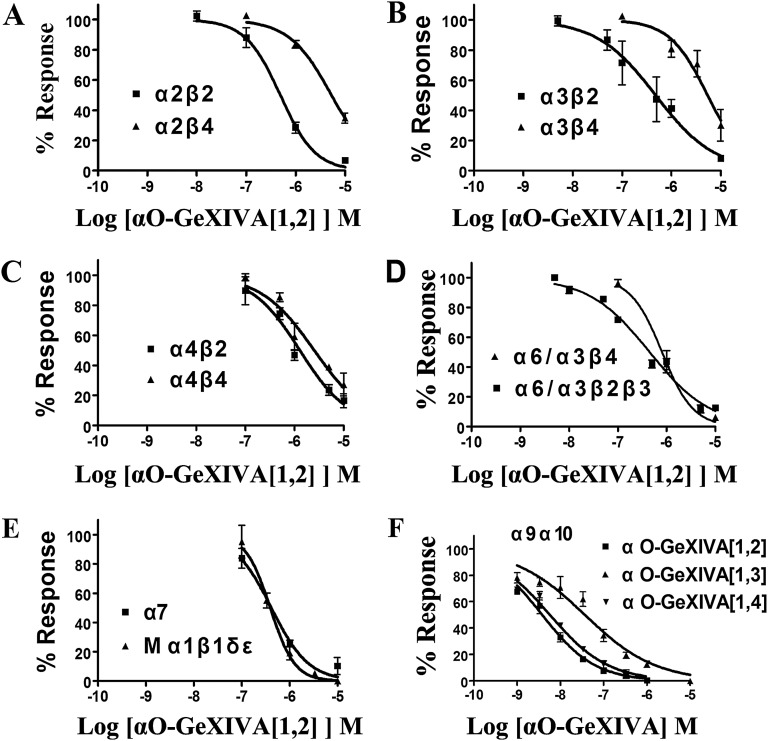
The effects of GeXIVA[1,2] on other nAChRs heterologously expressed in oocytes were tested. In contrast to its activity on α9α10 nAChRs, GeXIVA[1,2] was substantially less potent on the other nAChR subtypes (Fig. S2 A–E). For example, the IC50 ratios of each nAChR subtype compared with the IC50 ratio of α9α10 ranged from 86 (mouse muscle nAChR, α1β1δε) to over 1,100-fold (α3β4). The activities of GeXIVA[1,2] on rat α7 and mouse α1β1δε nAChRs were similar to each other, with IC50s of 415(264–655) and 394(311–498) nM, respectively. When a β4 rather than β2 nAChR subunit was coexpressed with the α2, α3, α4, and α6 subunits, the IC50 for GeXIVA[1,2] was much higher, suggesting that amino acid residue differences between the homologous β-subunits significantly influence GeXIVA[1,2] potency (Table 1).
Fig. S2.
Concentration responses of αO-conotoxin GeXIVA[1,2] on rat nAChR subtypes (A–E) and αO-conotoxin GeXIVA isomers on rat α9α10 nAChR in the presence of Ba++ (F). (A–E) IC50s and Hill slopes are shown in Table 2. Note that the peptide is generally more potent on β2- vs. β4-containing nAChRs. (F) Concentration response of equimolar Ba++ was substituted for Ca2+ in the perfusion solution, as described in Materials and Methods, to prevent activation of endogenous Xenopus oocyte Ca2+-activated Cl− currents. Values are mean ± SEM from six to nine separate oocytes. The IC50 for GeXIVA[1,2] was 3.8 nM (3.1–4.8 nM) with a Hill slope of 0.71 (0.58–0.84). The IC50 for GeXIVA[1,3] was 37 nM (25.0–55.7 nM) with a Hill slope of 0.54 (0.42–0.65). The IC50 for GeXIVA[1,4] was 5.8 nM (4.7–7.1 nM) with a Hill slope of 0.65 (0.56–0.73). The numbers inside the parentheses in the legend indicate 95% confidence interval of the IC50 or Hill slope.
Table 1.
IC50 and Hill slope values for blockade of various rat nAChR subtypes by αO-GeXIVA[1,2]
| Subtypes | IC50, nM* | Ratio† | Hill slope* |
| α9α10 | 4.61 (3.18–6.65) | 1 | 0.56 (0.44–0.69) |
| Mouse α1β1δε | 394 (311–498) | 85.5 | 1.71 (0.98–2.43) |
| α7 | 415 (264–655) | 90.0 | 1.12 (0.68–1.56) |
| α6/α3β2β3 | 450 (353–574) | 97.6 | 0.69 (0.59–0.79) |
| α6/α3β4 | 806 (664–980) | 173.7 | 1.36 (0.89–1.83) |
| α2β2 | 491 (352–684) | 106.5 | 1.23 (0.83–1.62) |
| α2β4 | 5,340 (4,200–6,790) | 1,158.4 | 1.02 (0.78–1.25) |
| α3β2 | 480 (275–837) | 104.1 | 0.73 (0.43–1.03) |
| α3β4 | 5,400 (3,480–8,410) | 1,171.4 | 1.13 (0.50–1.76) |
| α4β2 | 1,280 (953–1,720) | 277.7 | 0.88 (0.66–1.10) |
| α4β4 | 2,510 (1,630–3,870) | 544.5 | 0.79 (0.51–1.08) |
Numbers in parentheses are 95% confidence intervals.
nAChR subtype IC50/α9α10 IC50.
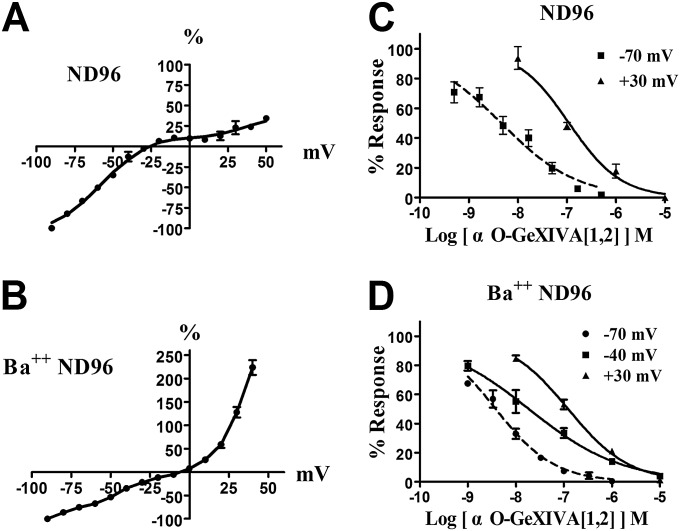
The α9α10 nAChRs are known to be highly permeable to calcium. In Xenopus oocytes, there is an endogenous Cl− channel and the entry of Ca2+ through the α9α10 nAChR elicits a secondary response via Ca2+-activated chloride currents. The magnitude of this response in Xenopus oocytes is large and can comprise >90% of the observed ACh-induced current (14, 15). We therefore wished to assess whether blockade of the ACh response by GeXIVA occurred by blockade of the α9α10 nAChR per se, or secondarily by blocking Ca2+-activated chloride currents. To accomplish this assessment, we substituted the closely related divalent cation Ba++, which does not elicit a chloride current, for Ca2+, and retested the effect on GeXIVA. Using the Ba++ buffer, the α9α10 nAChR was most potently blocked by GeXIVA[1,2] with an IC50 of 3.8 nM (3.1–4.8 nM); GeXIVA[1,3] and GeXIVA[1,4] had an IC50 of 37 nM (25.0–55.7 nM) and 5.8 nM (4.7–7.1 nM), respectively (Fig. S2F). Thus, the potencies of the GeXIVA isomers in the presence of Ba++ were similar to the potencies of the GeXIVA isomers seen in the presence of Ca2+, consistent with the toxin effect being due to blockade of the nAChR rather than blockade of Ca2+-activated chloride channels.
Mechanism of Blockade of α9α10 nAChR by GeXIVA[1,2].
We first investigated the voltage dependence of the blockade by GeXIVA[1,2]. Oocytes expressing α9α10 nAChR were voltage-clamped at holding membrane potentials ranging from −90 to +50 mV in ND96 and Ba++ ND96 media. Responses to ACh were calculated as the percentage of the response to ACh observed at −90 mV. Current–voltage relationships were determined by altering the voltage by 10-mV steps once every 1 min and measuring responses to a 1-s pulse of ACh (Fig. S3 A and B). Blockade by GeXVIA[1,2] was strongly voltage-dependent, and, consistent with previous reports, markedly more current passed through the channels at hyperpolarized potentials (15). The IC50 of GeXIVA[1,2] in ND96 at −70 mV was 24-fold lower than at +30 mV (Fig. S3C). Similarly, in Ba++ ND96, the IC50 at −70 mV of membrane potential was 32-fold lower than the IC50 of GeXIVA[1,2] in ND96 observed at +30 mV (Fig. S3D).
Fig. S3.
Voltage dependence of α9α10 nAChR blockade by αO-conotoxin GeXIVA[1,2]. Current responses were measured at membrane-holding potentials that were varied in 10-mV steps once per minute. Current–voltage responses were obtained in ND96 buffer (A) and buffer in which Ca2+ was replaced by Ba++ (Ba++ ND96) (B). Values were normalized to the response measured at −90 mV and represent the mean ± SEM. Concentration responses of αO-GeXIVA[1,2] on rat α9α10 nAChR at −70 mV and +30 mV in ND-96 (C) and at −70 mV, −40 mV, and +30 mV in Ba++ ND-96 (D) are shown. Values are mean ± SEM from five to 12 separate oocytes. In C, the IC50s in ND96 were −70 mV, 4.6 nM (3.2–6.7 nM) and +30 mV, 111 nM (73.8–166 nM). Hill slopes in ND96 were −70 mV, 0.56 (0.44–0.69) and +30 mV, 0.82 (0.56–1.08). In D, the IC50s in Ba++ ND96 were −70 mV, 3.8 nM (3.1–4.8 nM); −40 mV, 18.7 nM (1.7–29.9 nM); and +30 mV, 123 nM (100–151 nM). Hill slopes in Ba++ ND96 were −70 mV, 0.71 (0.58–0.84); −40 mV, 0.45 (0.35–0.55); and +30 mV, 0.68 (0.59–0.78). The numbers inside the parentheses in the legend indicate 95% confidence interval of the IC50 or Hill slope. Values are mean ± SEM.
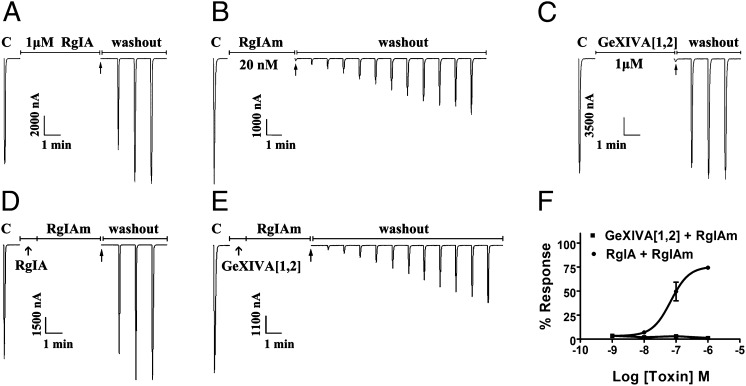
Voltage-dependent blockade suggests binding to an allosteric site or a channel pore of the receptor rather than the ACh-binding site. To assess the site of blockade, we used a different antagonist of the α9α10 nAChR, RgIAm. RgIAm [sequence GCCTDPRCCitTyr(I)QCR, where Cit is citrulline and Tyr(I) is monoiodo-Tyr] is an analog of α-conotoxin RgIA, a competitive inhibitor of α9α10 nAChRs (16, 17). RgIAm was chosen for these experiments because blockade by RgIAm is slowly reversed upon toxin washout, whereas the block by either GeXIVA[1,2] or RgIA was rapidly reversed (Fig. 3). We reasoned that if the toxin-binding sites overlap, GeXIVA[1,2] should prevent binding of RgIAm to the α9α10 nAChR when both toxins are present in solution. However, preincubation with a high concentration of GeXIVA[1,2] failed to prevent subsequent access to the nAChR by RgIAm, as evidenced by the slow recovery from washout of both toxins. As a control, we repeated the experiment using native RgIA (which has rapid disassociation kinetics) in place of GeXIVA[1,2]. In contrast to the result with GeXIVA[1,2], preincubation with RgIA prevents subsequent binding of RgIAm, consistent with binding to the same site on the receptor. Thus, GeXIVA[1,2] and RgIAm appear to define distinct, nonoverlapping binding sites on the α9α10 nAChR.
Fig. 3.
Binding site comparison of αO-GeXIVA and α-conotoxin RgIAm. In each instance, toxin solution was applied to α9α10 nAChRs expressed in Xenopus oocytes, followed by toxin washout. The response to 1-s pulses to ACh were measured as described in Materials and Methods. (A) One micromolar α-conotoxin RgIA for 5 min. Note the rapid recovery following toxin washout. (B) Twenty nanomolar α-conotoxin RgIAm for 4 min. Note the slow recovery. (C) One micromolar αO-conotoxin GeXIVA[1,2] for 5 min. Note the rapid recovery. (D) One micromolar α-conotoxin RgIA for 1 min, followed by 20 nM α-conotoxin RgIAm for 4 min. Note that preblockade of α9α10 nAChR with α-conotoxin RgIA prevented the slowly reversible block associated with α-conotoxin RgIAm, consistent with competitive binding to the same or overlapping site. (E) One micromolar αO-GeXIVA[1,2] for 1 min, followed by 20 nM α-conotoxin RgIAm for 4 min. Note that preblockade of α9α10 nAChR with αO-GeXIVA[1,2] did not prevent the very slowly reversible block associated with α-conotoxin RgIAm, consistent with occupancy of distinct binding sites. (F) Oocytes expressing α9α10 nAChRs were preincubated with varying concentrations of RgIA for 5 min. This step was followed by coincubation with RgIA + 20 nM RgIAm for a further 5 min. Both toxins were then washed out, and the response was measured following 2 min of washout. Note that an increasing concentration of RgIA led to a greater percent response (faster recovery) to ACh. Oocytes expressing α9α10 nAChRs were preincubated with varying concentrations of GeXIVA[1,2] for 5 min. This step was followed by coincubation with GeXIVA[1,2] + 20 nM RgIAm for a further 5 min. Both toxins were then washed out, and the response was measured following 2 min of washout. Note that an increasing concentration of GeXIVA[1,2] had no effect on percent response to ACh (n = 5–6 oocytes).
We then tested the concentration dependence of the effects of preincubation of RgIA or GeXIVA[1,2] on the washout kinetics of blockade by 20 nM RgIAm. Blockade of α9α10 nAChRs by either RgIA or GeXIVA[1,2] is reversed after 2 min of toxin washout. In contrast, after application of RgIAm, >95% of the α9α10 nAChR response remains blocked 2 min after RgIAm washout. We therefore preincubated the oocytes with varying concentrations of RgIA or GeXIVA[1,2], followed by coincubation of RgIA + 20 nM RgIAm or GeXIVA[1,2] + 20 nM RgIAm, respectively. The percentage of the block remaining after a 2-min of washout of both toxins was then assessed. Preincubation with RgIA followed by coapplication of RgIA + RgIAm resulted in an RgIA concentration-dependent effect on recovery of blockade. Increasing concentrations of the rapidly reversible RgIA resulted in faster recovery from nAChR blockade by RgIA + RgIAm, and therefore less total blockade 2 min after toxin washout (Fig. 3F). This result is consistent with RgIA preventing the subsequent binding of RgIAm. In contrast, increasing concentrations of GeXIVA[1,2] had no effect on the recovery from blockade by GeXIVA[1,2] + RgIAm at any GeXIVA[1,2] concentration tested. This result is consistent with GeXIVA[1,2] binding to a separate site from RgIAm.
Lack of Effect of GeXIVA on Voltage-Dependent Calcium Channels.
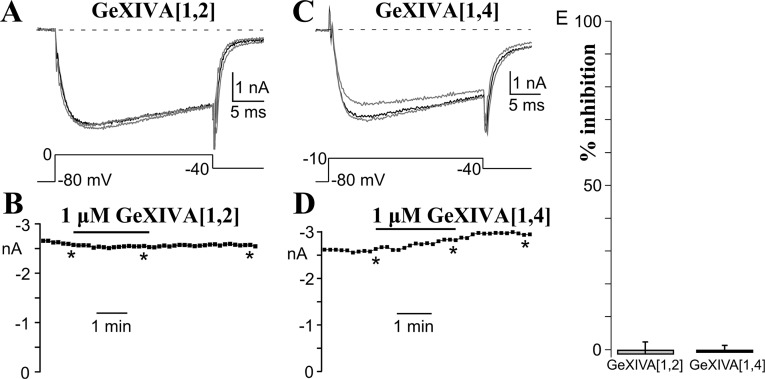
Previously identified members of the conotoxin O-superfamily are potent blockers of VGCCs. We therefore tested the effects of GeXIVA isomers on VGCCs from dissociated rat dorsal root ganglia (DRG) neurons. Neither GeXIVA[1,2] nor GeXIVA[1,4] at 1 μM significantly affected calcium currents in DRG neurons (Fig. S4). DRG calcium channels are composed of ∼50% N type (CaV2.2), ∼24% P/Q type (CaV2.1), and ∼13% L type (CaV1.2), with the remainder being R type (probably CaV2.3) (18–20).
Fig. S4.
αO-contoxins GeXIVA[1,2] and GeXIVA[1,4] do not affect voltage-dependent calcium current in rat DRG neurons. Different DRG neurons were exposed for 2.5 min to 1 μM of either GeXIVA[1,2] (A) or GeXIVA[1,4] (C). Currents were elicited at the indicated voltages (Materials and Methods) once every 10 s, and are shown before (gray trace), during (black trace), and upon (gray trace) recovery from toxin. The charge carrier was 5 mM Ba2+. (B and D) Time course is shown for the data depicted above. The asterisks indicate the times when the currents shown above were generated. The solid bar indicates the duration of toxin application. (E) Average effect of toxin on voltage-dependent calcium current (n = 8, 8). The duration of toxin application ranged from 1.2 to 3.1 min for GeXIVA[1,2] (mean = 1.7 min) and from 1.5 to 2.5 min for GeXIVA[1,4] (mean = 1.9 min). Neither toxin significantly affected calcium current (P > 0.05). The neuronal diameter ranged from 21 to 34 μm (mean = 27 μm) for the GeXIVA[1,2]-exposed cells and from 26 to 32 μm (mean = 30 μm) for the GeXIVA[1,4]-exposed cells.
Structural Characterization of GeXIVA.
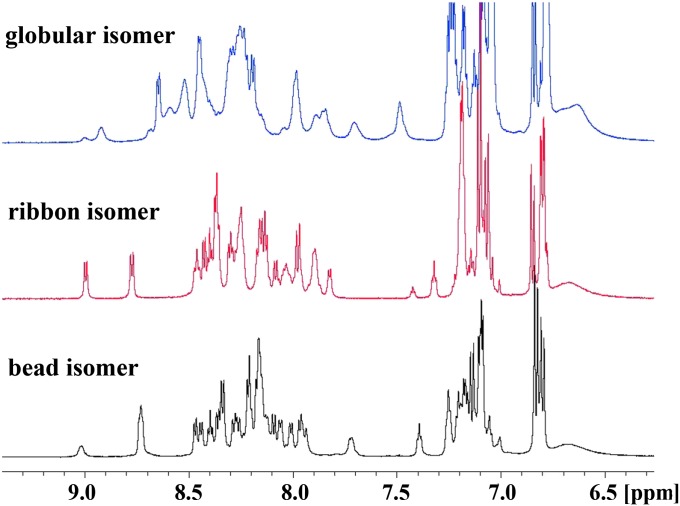
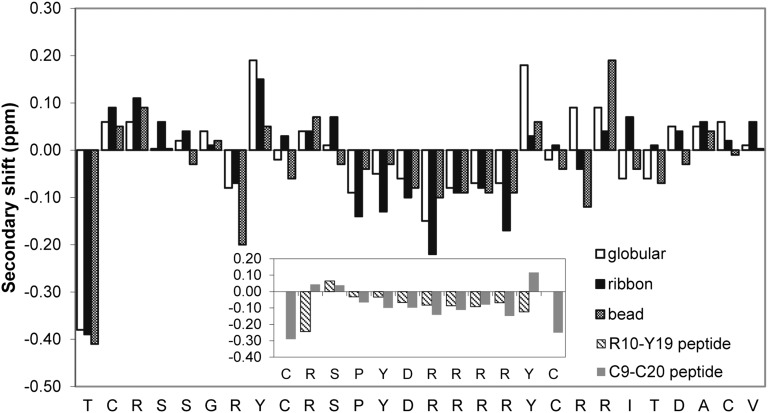
The three disulfide isomers of GeXIVA were examined by NMR to determine their solution structures. Unexpectedly, the 1D NMR spectrum of the globular isomer displayed significant peak broadening, potentially indicative of either conformational exchange or aggregation, but the latter was eliminated by the results of NMR diffusion experiments, which suggested a hydrodynamic radius consistent with a monomer. The 1D spectra of the ribbon and bead isomers were much sharper (Fig. 4). The 2D NMR spectra of the bead and ribbon isomers were fully assigned, but not the 2D NMR spectra of the globular isomer due to peak overlapping and broad lines. Changes in temperature and pH did not resolve this overlap. Analysis of αH secondary shifts, calculated by subtracting random coil αH shifts (21) from the experimental Hα shifts, showed little evidence of well-defined secondary structure elements in any of the isomers (Fig. S5). The secondary shifts in the middle of the peptide sequence (residues 9–20) suggest a tendency toward helical content, particularly for the ribbon isomer, but addition of helix-inducing d3-trifluoroethanol (TFE) as a cosolvent caused little change to the secondary αH shifts. Fig. S6 (Inset) shows the secondary αH shifts of two synthetic peptides corresponding to the “loop” region of the ribbon isomer, or the “linker” region of the bead isomer. The data indicate that the short linear peptide (from R10–Y19) is essentially a random coil when removed from the larger peptide framework; however, inclusion of a disulfide bond bridging its termini induces a helical nature, as evidenced by slightly increased secondary shifts.
Fig. 4.
Amide region of the 1H-NMR spectra of GeXIVA recorded at 298 K and 600 MHz. Globular, GeXIVA[1,3]; ribbon, GeXIVA[1,4]; and bead, GeXIVA[1,2] isomers are shown.
Fig. S5.
Secondary αH shifts of the three isomers of GeXIVA (globular, ribbon, and bead) in water. (Inset) Secondary αH shifts of synthetic peptides corresponding to residues from R10–Y19 and from C9–C20.
Fig. S6.
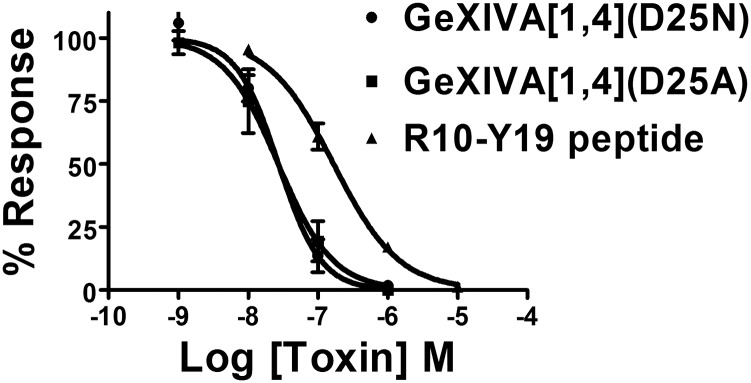
Concentration response curves of GeXIVA analogs. Compounds were tested on α9α10 nAChRs expressed in Xenopus oocytes. The peptide with sequence RSPYDRRRY had an IC50 of 168 (129–217) nM, with a Hill slope of 0.93 (0.71–1.1). GeXIVA[1,4](D25N) had an IC50 of 27 (18–42) nM, with a Hill slope of 1.4 (0.89–2.0). GeXIVA[1,4](D25A) had an IC50 of 27 (15–50) nM, with a Hill slope of 1.1 (0.53–1.7). The numbers inside the parentheses in the legend indicate 95% confidence interval of the IC50 or Hill slope. Values are mean ± SEM (n = 3–6 oocytes per data point).
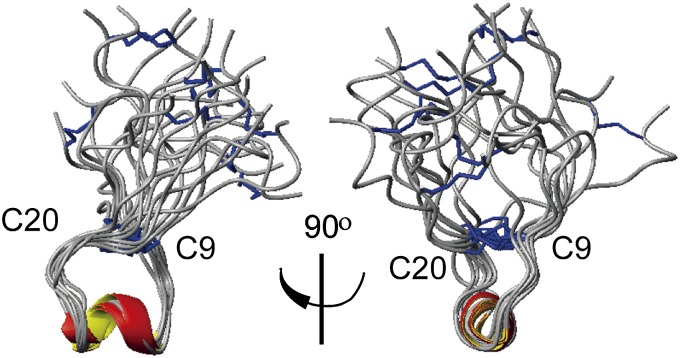
Structure calculations were performed only on the ribbon and bead isomers because the globular form apparently does not have a single conformation. Because the secondary shifts suggested a slightly better defined structure for the ribbon isomer, additional 900-MHz NMR data were acquired for this peptide. Nevertheless, neither ribbon nor bead isomer gave a precisely overlaid ensemble of structures when the superposition was made across the entire molecule. However, closer inspection of the disulfide-restrained loops at either end of the bead isomer shows that these loops (C2–C9 and C20–C27) are internally well defined (Fig. 1 and Table S2) but are “uncoupled” from one another by the 10–19 linker region, which has a slight helical tendency. The larger loop between C9 and C20 of the ribbon isomer contains a better defined but short 310-helix (from Y12–R16), and this helix–loop region overlays well between NMR models (Fig. 5; rmsd of 0.99 ± 0.33 Å across backbone atoms).
Table S2.
Experimental restraints and structural statistics for GeXIVA, bead, and ribbon
| Restraints and statistics | Ribbon | Bead |
| Experimental data | ||
| Distance restraints | ||
| Total NOE | 155 | 234 |
| Intraresidue | 87 | 99 |
| Interresidue | 68 | 134 |
| Sequential (|I − j|) | 54 | 111 |
| Medium-range (|I − j| ≤ 4) | 14 | 23 |
| Long-range (|I − j| ≥ 5) | 0 | 0 |
| Dihedral angle restraints, ϕ | 43 | 40 |
| Total NOE violations exceeding 0.3 Å | 0 | 0 |
| Total dihedral violations exceeding 2.0° | 0 | 0 |
| Structure statistics | ||
| Average pairwise rmsd, Å | ||
| Backbone (heavy) atoms | 3.80 ± 1.44 (5.27 ± 1.90) | 5.1 ± 1.3 (6.5 ± 1.5) |
| Backbone (heavy) atoms (residues 9–20) | 0.99 ± 0.33 (2.84 ± 0.82) | |
| Backbone (heavy) atoms (residues 2–9) | 0.76 ± 0.25 (2.16 ± 0.57) | |
| Backbone (heavy) atoms (residues 20–27) | 1.66 ± 0.60 (3.44 ± 0.96) | |
| Ramachandran statistics* | ||
| Most favored and allowed regions, % | 71.7 | 63.3 |
| Generously allowed regions, % | 28.3 | 36.7 |
| Disallowed regions, % | 0 | 0 |
As analyzed by PROCHECK-NMR.
Fig. 5.
Ribbon diagram of the NMR solution structure of GeXIVA[1,4]. Images are overlaid across the C9–C20 loop. Disulfide bonds are depicted in blue. α-Helical regions are colored red and yellow.
Molecular Modeling of GeXIVA Interactions.
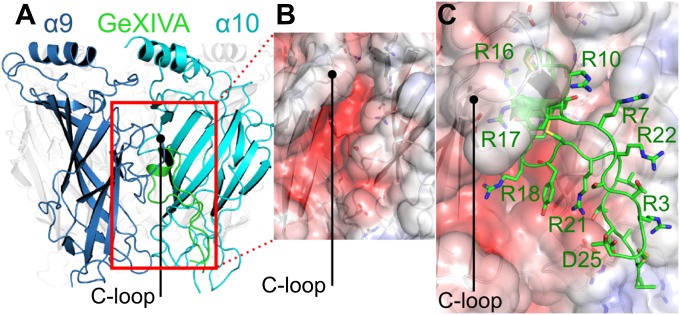
A potential binding site of GeXIVA located at the interface between the α9 (principal) and α10 (complementary) subunits was suggested by molecular modeling. However, it has been shown (16) that the major contribution to the high-affinity binding of RgIA comes from the complementary (−) side of the α9 subunit. The noncompetitive interaction of GeXIVA with RgIA suggests that it is not engaged in the interaction with the α10(+)/α9(−) interface, and GeXIVA could therefore interact either in the α9α10 pocket or at other sites, such as an allosteric binding site. GeXIVA bears nine positively charged residues, suggesting that electrostatic interactions should be important at the interface. The bead and ribbon isomers have similar binding affinities for α9α10, suggesting that they interact using nonspecific interactions involving long range electrostatic interactions or that the two isomers share a similar local structure crucial for interacting with the receptor. NMR analysis showed that the central part of the peptide, from positions 12–18, is helical for the ribbon isomer and has a small helical propensity for the bead isomer. This shared helical motif between the two isomers is reminiscent of the helix displayed by framework I α-conotoxins (22), most of which seem to adopt a conserved binding mode, with their α-helix deeply buried in the ligand-binding pocket. This pocket is located between the C-loop of the principal subunit and a β-sheet from the complementary subunit (Fig. 6A).
Fig. 6.
Model of the interaction between GeXIVA[1,4] and the α9α10 subunit orthosteric binding site located between α9 (principal) and α10 (complementary) subunits. (A) Binding site is mainly contributed by the C-loop of the α9 subunit and a β-sheet from the α10 subunit. The α-helix of GeXIVA binds at the same location as framework I α-conotoxin but in a reverse orientation. (B) Electrostatic potential generated by the α9α10 ligand-binding domain mapped on the solvent-accessible surface of the GeXIVA-binding site. The solvent-accessible surface is colored from red to blue corresponding to electrostatic potentials from −5 kT/e and below to +5 kT/e and above, respectively. The binding site creates a negative charge potential compatible with the binding of GeXIVA, which is highly positively charged. (C) GeXIVA is tightly packed in the binding site, and GeXIVA-charged side chains R16 and R17 establish salt bridges deep in the pocket in the model. The molecular model was built by homology with the crystal structure of AChBP in complex with conotoxin [A10L,D14k]PnIA [Protein Data Bank (PDB) ID code 2BR8], as well as the extracellular domain of the monomer α1 subunit (PDB ID code 2QC1). The model was refined by 15-ns molecular dynamics simulations.
A model of the α9α10 orthosteric binding site, in which the α9 is the principal subunit, shows that it is highly electronegative (Fig. 6B), suggesting that it could be the binding site of the positively charged GeXIVA. A model of GeXIVA docked into this pocket was generated by positioning the helix of the ribbon isomer at the same location as the location of the framework I α-conotoxins. Of the two possible orientations of the helix, one has good complementarity of charges, whereas the other would create repulsive interactions between positive charges of Arg16, Arg17, and Arg18 of GeXIVA and Arg111 located proximate to the binding site on the α10 subunit. The model of the best orientation was stable over 15-ns molecular dynamics simulations, and displays no distortion of the C-loop, a stable helix, good packing, and charge complementarity (Fig. 6C). The termini of GeXIVA protrude out of the pocket, suggesting that the helix of the bead isomer should be able to dock similarly.
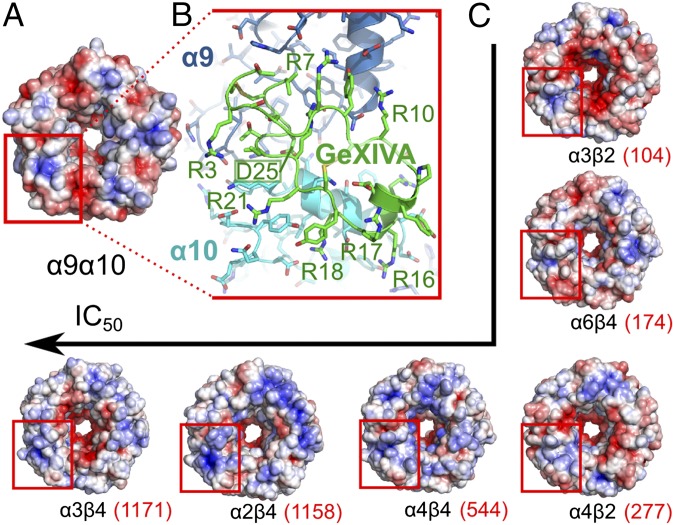
An interaction in the α9α10 orthosteric ligand-binding pocket cannot explain why the block of α9α10 by GeXIVA is voltage-dependent, but a second GeXIVA-binding site on α9α10, possibly having an allosteric effect, was predicted to exist in proximity to the pore using further molecular modeling. Putative allosteric sites for GeXIVA were searched using five coarse-grained molecular dynamics simulations of the α9α10 ligand-binding domain and ribbon GeXIVA. Two simulations showing binding on the membrane side of the pore were discarded because this site is not accessible in the full-length receptor. In one simulation, GeXIVA interacted below the α9α10 orthosteric binding site, but this binding mode was not completely stabilized after a further 200-ns simulation, indicating that it is short-lived, possibly leading to a transition toward binding in the orthosteric site. Finally, the two other simulations resulted in binding at the outskirts of the pore between the α9 and α10 subunits. Of these two simulations, the one with the higher stability displayed a binding mode in which Asp25 of GeXIVA is buried in a small, positively charged pocket between the two subunits and all of the Arg of GeXIVA forms an extended number of electrostatic interactions with negatively charged receptor side chains located on both the α9 and α10 subunits (Fig. 7 A and B). As shown in Fig. 7C, other nAChR subtypes that GeXIVA blocks with high nanomolar IC50 display a similar electropositive pocket surrounded by negatively charged residues, but the electronegative patch seems to be absent from the subtypes blocked by GeXIVA at micromolar range IC50. The two binding sites that were suggested by molecular modeling can potentially correspond to low- and high-affinity binding sites, which have been shown to exist for other α-conotoxins (23). To test these predictions, we synthesized three mutants of GeXIVA. First, we synthesized a truncated version of GeXIVA containing the central residues R10–Y19. This nine-residue peptide potently blocked the α9α10 nAChR with an IC50 of 178 nM (Fig. S6), consistent with interaction of this segment of GeXIVA binding to the receptor.
Fig. 7.
Potential second binding site of GeXIVA [1,4] on the extracellular domain of α9α10 and other nAChRs. (A) α9α10 Subtype displays an electropositive pocket surrounded by two negatively charged patches next to the central pore on the ligand-binding domain. (B) This patch was identified by coarse-grained molecular docking as a potential interaction site of GeXIVA, with its side chain, D25, occupying the positively charged pocket and several of its Arg side chains establishing charges and salt bridge interactions with positively charged residues of α9 and α10 subunits. (C) Same motif of electrostatic potential could be identified on the surface of other tested nAChR subtypes, and the integrity of this motif seems to correlate with the activity of GeXIVA. The fold difference from the activity on α9α10 is given in parentheses. The molecular models of the nAChR ligand-binding domains were built by homology with AChBP structure (PDB ID code 2BR8), the extracellular domain of monomer α1 subunit (PDB ID code 2QC1), and the structure of the β-subunit in the EM structure of Torpedo marmorata muscle type nAChR (PDB ID code 2BG9). The solvent-accessible surfaces were colored from red to blue, corresponding to electrostatic potentials from −5 kT/e and below to +5 kT/e and above, respectively.
Next, we mutated Asp25 to either Asn (GeXIVA[1,4](D25N)) or Ala (GeXIVA[1,4](D25A)). Both peptides had an IC50 of 27 nM, which was three- to fourfold higher than the parent peptide (Fig. S6). Thus, replacement of the negatively charged Asp with a neutral residue reduced potency, consistent with the model.
Effects of GeXIVA[1,2] on Chronic Constriction Injury-Induced Mechanical Hyperalgesia.
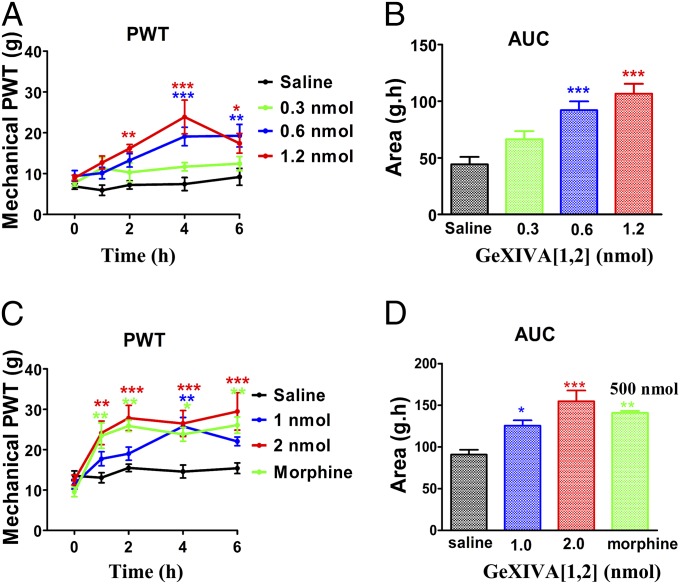
Constriction of the sciatic nerve is a commonly used model of chronic neuropathic pain (24, 25). Postinjury thresholds to mechanical stimuli were evaluated 11–13 d after surgery. Eligible chronic constriction injury (CCI) rats were treated with an i.m. injection of saline alone, 0.3–2 nmol of GeXIVA[1,2], or 500 nmol of morphine, which served as a positive control. Preinjection paw-withdrawal threshold (PWT) (t = 0) and post-CCI PWT 1–6 h postinjection were then assessed. GeXIVA[1,2] significantly reduced mechanical hyperalgesia induced by CCI (Fig. 8). The GeXIVA[1,2]-treated animals showed significantly increased PWTs relative to pre-CCI (t = 0) and relative to the saline-treated group (Fig. 8 A and C). The robust increase in PWT persisted at least 6 h after injection of GeXIVA[1,2]. The area under the curve (AUC) was calculated for each treatment; GeXIVA[1,2] displayed a significant increase in AUC as a function of increasing dose. Doses of 1 and 2 nmol produced effects that did not differ significantly from the effect of 500 nmol of morphine (P > 0.05).
Fig. 8.
Effect of αO-conotoxin GeXIVA[1,2] on mechanical hyperalgesia. The sciatic nerve of rats was loosely ligated to produce CCI, a model of human neuropathic pain, as described in Materials and Methods. Rats were injected i.m. with 0.9% saline, conotoxin, or morphine. PWT was used as a measure of mechanical hyperalgesia. (A) Effect of compound vs. time was assessed using a series of von Frey hairs. Time point t = 0 represents mechanical PWT immediately before injection. (B) Corresponding dose–response calculated as AUC for data from each dose of drug in A for time points between 0 and 6 h. (C) PWT was assessed for a separate group of rats using an electronic von Frey anesthesia meter. (D) AUC for data from each dose of drug in C. (n = 8 for A and B; n = 6 for C and D.) Each point in A and C represents the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; Dunnett’s post hoc tests.
Effect of GeXIVA[1,2] on Motor Function.
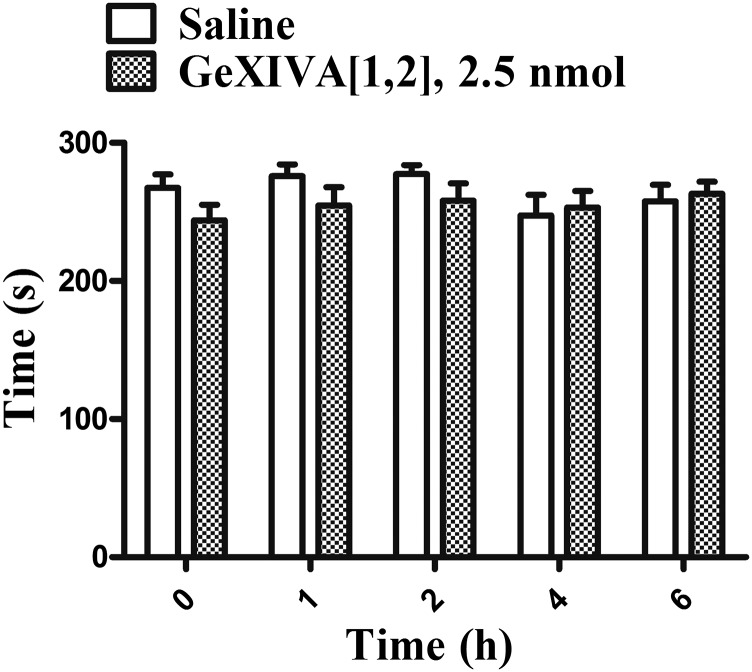
Potential effects of 2.5 nmol of GeXIVA[1,2] on rotarod latencies (n = 8) were measured to assess whether the conotoxin caused motor impairment in rats. GeXIVA[1,2] had no significant effect on the performance of rats in the accelerating rotarod test during any of the time points 1, 2, 4, or 6 h after i.m. injection (Fig. S7). Thus, motor performance was not impaired and did not interfere with hyperalgesia testing.
Fig. S7.
Effect of the peptide αO-conotoxin GeXIVA[1,2] on motor performance. Each bar represents the mean ± SEM change in rotarod latency (s) of preinjection (t = 0) and at 1 h, 2 h, 4 h, and 6 h after injection with saline or peptide (2.5 nmol) (n = 8).
Discussion
In this study, we characterized a previously unidentified peptide GeXIVA from the South China Sea mollusk C. generalis. Homology in the pre-propeptide sequence indicates that it is a member of the O1 gene superfamily. However, to our knowledge, GeXIVA is the first member of this superfamily with four rather than six Cys residues, and this structural difference translates into striking functional differences. Whereas many other O-superfamily peptides block voltage-gated Ca2+ channels, GeXIVA is inactive at these targets. The ω-conotoxins were the first members of the O-superfamily to be identified, and they potently block various subtypes of VGCCs, with pharmaceutical applications in some cases. For example, the selectivity of ω-conotoxin MVIIA for N-type calcium channels has led to its use as an intrathecally delivered drug to treat refractory chronic pain in patients with cancer (26). It is remarkable that the newly characterized peptide is inactive at VGCCs but blocks nAChRs, most potently the α9α10 nAChR subtype.
The α9α10 nAChR has been implicated in pain signaling, and α-conotoxins that block this receptor are effective in the chronic constriction nerve injury model of neuropathic pain (27). Therefore, GeXIVA was tested using the chronic constriction nerve injury model and shown to reduce mechanical hyperalgesia dose-dependently from CCI of the sciatic nerve. Motor performance was not impaired, indicating that analgesic effects were not confounded by motor deficits. Analgesic effects occurred at low doses (2 nmol or lower), with the 2-nmol dose producing effects greater than or equal to a dose of 500 nmol of the opioid analgesic morphine. Several studies have shown that some α-conotoxins block VGCCs via stimulation of GABAB receptors, suggesting that antinociceptive effects were mediated through blockade of N-type calcium channels (28–33). However, some studies have not fully reproduced this effect (34, 35), and we show that GeXIVA fails to block VGCCs. Because GeXIVA is active at α9α10 nAChRs, but not VGCCs, our findings provide support for the involvement of α9α10 nAChRs in pain sensation. GeXIVA is active after i.m. injection, rather than requiring intrathecal administration like the analgesic ω-conotoxin MVIIA that targets Ca2+ channels. The size and positive charge of GeXIVA make it unlikely that it would cross the blood–brain barrier, suggesting a peripheral site of action.
α-Conotoxins are competitive blockers of nAChRs, and their blocking of α9α10 nAChRs occurs by occupation of a site at the α10/α9 subunit interface (16, 33, 36). There are no characterized radioligands for α9α10 nAChRs or cell lines that robustly express α9α10 nAChRs to perform radioligand competition binding assays. Functional competition experiments with α9α10 nAChRs expressed in oocytes suggested that GeXIVA and RgIAm may bind to separate sites. In addition, the block of α9α10 nAChR by GeXIVA is strongly voltage-dependent, consistent with noncompetitive binding to α9α10 nAChR. Thus, αO-conotoxin GeXIVA represents a previously uncharacterized probe that may further define binding sites on the α9α10 nAChR. Its three active disulfide isomers provide additional opportunities for elucidating structure–activity relationships. Interestingly, the globular isomer was the least potent of the three. Whereas there is precedent for high potency of ribbon isomers (e.g., for α-conotoxin AuIB) (37), high potency of a bead form is rare. However, bead isomers have been infrequently studied, and our findings hint that greater attention to these isomers might lead to a fuller understanding of conotoxin structure–activity relationships.
The characteristics of GeXIVA are compared with other selected conotoxins in Table 2. Most O1-superfamily conotoxin mature peptide regions share a conserved arrangement of six Cys residues (C-C-CC-C-C, framework VI/VII). These include the well-known conotoxins that block VGCCs. In addition, precursor peptides highly homologous to GeXIVA have been cloned from C. generalis and Clonus miles. The function of these two peptides is unknown, but their precursors differ in their mature toxin regions by having five and six Cys, respectively (38, 39), instead of the four Cys found in GeXIVA.
Table 2.
Comparison of αO-GeXIVA with selected conotoxins
| Toxin | Source | Sequence | Target | Mass, Da | Source |
| αO-GeXIVA | C. generalis | TCRSSGRYCRSPYDRRRRYCRRITDACV* | α9α10 nAChR >> α1β1δε ∼ α7 ∼ α*β2 nAChRs > α*β4 nAChRs | 3,452 | This study |
| αB-VxXXIVA | C. vexillum | VRCLEKSGAQPNKLFRPPCCQKGPSFARHSRCVYYTQSRE* | α9α10 nAChR > Mouse α1β1γδ | 4,623 | (71) |
| Conantokin-Br (con-Br) | C. brettinghami | GDγγYSKFIγRERγAGRLDLSKFP* | NMDAR, NR2B > NR2D > NR2A > NR2C | 3,046 | (72) |
| Mr8.2 | C. marmoreus | ALVSQCRPCPTCRECKCRECKCRECQCRIHSCLSAWDSRGIWMRT | ND | 5,280 | (73) |
| ω-MVIIA | C. magus | CKGKGAKCSRLMYDCCTGSCRSGKC† | Cav2.2 > Cav2.1 | 2,637 | (74, 75) |
| pl14a (κJ-PlXIVA) | C. planorbis | FPRPRICNLACRAGIGHKYPFCHCR† | kV 1.6 > 1.1 > 1.2 ∼ 1.3 ∼ 1.4 ∼ 1.5 ∼ 2.1 ∼ 3.4 | 2,909 | (76, 77) |
| pl14a (αJ-PlXIVA) | Muscle > α3β2 nAChRs | ||||
| lt14a (αL-LtXIVA) | C. litteratus | MCPPLCKPSCTNC† | ND (inhibits contraction of the frog muscle) | 1,391 | (78) |
| vil14a (κL-VilXIVA) | C. villepinii | GGLGRCIYNCMNSGGGLSFIQCKTMCY* | ND | 2,871 | (40) |
| ψ-PIIIE | C. purpurascens | HOOCCLYGKCRRYOGCSSASCCQR† | Muscle nAChR | 2,715 | (79, 80) |
| α-LtIA | C. litteratus | GCCARAACAGIHQELC† | α3β2 > α6/α3β2β3 | 1,600 | (81) |
| αA-EIVA | C. ermineus | GCCGPYONAACHOCGCKVGROOYCDROSGG† | Muscle nAChR | 3,094 | (82) |
| αC-PrXA | C. parius | TYGIYDAKPOFSCAGLRGGCVLPONLROKFKE† | Muscle nAChR | 3,539 | (83) |
| αD-VxXXB | C. vexillum | DDγSγCIINTRDSPWGRCCRTRMCGSMCCPRNGCTCVYHWRRGHGCSCPG (dimer) | α7 > α3β2 | 5,735 | (84) |
| αS-RVIIIA | C. radiatus | KCNFDKCKGTGVYNCGγSCSCγGLHSCRCTYNIGSMKSGCACICTYY | Muscle nAChR | 5,167 | (85) |
| Neuronal nAChR |
C., Conus; ND, not determined; O, hydroxyproline; γ, γ-carboxyglutamate (Gla).
C terminus COOH.
C terminus amidation.
In the current study, three different disulfide isomers of GeXIVA were synthesized and assessed functionally and structurally. The globular isomer has two or more conformations that coexist in intermediate exchange on the NMR time scale. In contrast, the ribbon and bead isomers have sharper spectra indicative of either single conformations or fast exchange between conformations. The ribbon isomer contains a well-defined loop of 12 residues restrained by a disulfide bond and featuring a 310-helix between Tyr12 and Arg16.
It is interesting to note the differences between GeXIVA and other reported framework 14 conotoxins, particularly the loop size separating the disulfide bonds. In each of these conotoxins, at least two of the loops are small (i.e., one to three residues), whereas a third loop is often much larger (10–11 residues). For example, both Vil14a and Flf14a share the pattern of C(X3)C(X11)C(X3)C (40), whereas Pu14a has the pattern C(X10)C(X)C(X3)C. In contrast, GeXIVA has three loops of 6, 10, and 6 residues. These three large loops probably contribute to the flexibility of the globular isomer and to the ill-defined nature of the midloop of the bead isomer. Among the ∼80 known framework XIV conopeptides, GeXIVA stands out as having the largest number of Arg residues, which have been suggested from the modeling studies to be crucial for inhibitory activity at the α9α10 nAChR subtype. The nAChRs are found at neuromuscular and/or neuroneuronal synapses, as well as at extrasynaptic locations across the whole phylum of bilateria from nematodes to mammals (41). Previously characterized conotoxin nAChR antagonists have been members of the A-, B-, C-, D-, M-, S-, L- or J-superfamily of conotoxins. Overall, there are eight different families of conotoxins that are known to target the nAChRs: α-conotoxins, αB-conotoxins, αC-conotoxins, αD-conotoxins, ψ-conotoxins, αS-conotoxins, αL-conotoxins, and αJ-conotoxins (Table 2). GeXIVA is the first peptide with potent activity at the nAChRs that belongs to the O1-gene superfamily. Its structure provides another illustration of the diverse range of peptides from cone snails that target nAChRs. Only 10% of known conopeptides have been at least partly functionally characterized, ca. 200 of ca. 2,000 conopeptides, and it has been estimated that the total number of distinct conopeptides could reach hundreds of thousands (42). The previously unidentified activity displayed by GeXIVA, which blocks nAChR differently from framework I-conotoxins, suggests that a wide range of conopeptides with yet other novel activities on the nervous system await discovery.
The α9α10 nAChR is of increasing interest in biomedicine, and α-conotoxins that block the α9α10 nAChR subtype are analgesic (9, 27, 43–45). The 4.6 nM IC50 of GeXIVA[1,2] places it among the most potent peptides that target α9α10 nAChRs; the IC50s (nanomolar) for α-conotoxins Vc1.1, RgIA, and PeIA are 19, 5.2, and 6.9, respectively (43, 46). Small-molecule antagonists of α9α10 nAChRs (which have IC50s as low as 0.51 nM) are also analgesic in rodent models of neuropathic pain (47–49), further validating α9α10 nAChR as an important therapeutic target. Each of these tested α9α10 antagonists produces long-lasting analgesia after single injection. The α-conotoxins show an increasing effect after repeated administration, suggesting a possible disease-modifying effect (9, 27, 43). Similar testing of GeXIVA, as well as head-to-head comparisons with these other α9α10 nAChR antagonists, will be of interest. Development and testing of selective α9α10 antibodies in pain models would also be of interest for future studies. Separately, blockade of the α9α10 nAChR has been shown to slow the development of breast cancer in animal models, and these receptors may also be therapeutically relevant to lung cancer (50–52). Novel antagonists of the α9α10 nAChR, such as GeXIVA, represent valuable probes of this receptor subtype and could be considered for the development of new drug leads.
Materials and Methods
Materials.
C. generalis specimens were collected from the South China Sea off Hainan Province. Venom ducts were frozen and stored at −80 °C.
cDNA Cloning and Sequencing.
Total RNA was extracted from C. generalis venom ducts and purified using an RNA isolation kit, which was described previously (53). cDNA was prepared by RT of the total RNA. The resulting cDNA served as a template for RT-PCR, which was performed using primers on conserved elements in the UTRs of O1- superfamily conotoxins as follows: forward primer 1, 5′ CATCGTCAAGATGAAACTGACGTG 3′ (24 bp); reverse primer 2, 5′ CACAGGT ATGGATGACTCAGG 3′ (21 bp). Then, the online ProP 1.0 Server was used to predict the signal and mature peptide of the conotoxin precursors (54).
Peptide Synthesis.
The peptide sequence was assembled by solid-phase methodology on an ABI 433A peptide synthesizer (Applied Biosystems) using Fmoc chemistry and standard side-chain protection, except for Cys residues. Cys residues of the three possible isomers were protected in pairs with either S-trityl on Cys20 and Cys27 (designated GeXIVA[1,2]), Cys9 and Cys27 (designated GeXIVA[1,3]), or Cys9 and Cys20 (designated GeXIVA[1,4]) or S-acetamidomethyl on Cys2 and Cys9, Cys2 and Cys20, and Cys2 and Cys27. A two-step oxidation protocol was used to fold the peptides as described previously (55). α-Conotoxin RgIA and its analog, RgIAm, were synthesized as previously described (16, 17).
cRNA Preparation and Injection.
Oocytes were injected within 1 d of harvesting, and recordings were made 1–5 d postinjection.
Voltage-Clamp Recording.
Oocytes were voltage-clamped at −70 mV at room temperature. In addition, a range of membrane potentials from −90 mV to +50 mV was tested for rat α9α10 nAChR (15). The activity of the most potent isomer of GeXIVA was tested on rat α9α10 nAChR at +30 mV and −40 mV. Oocytes were voltage-clamped and exposed to ACh and peptide as described previously (56).
Voltage-Dependent Calcium Current Recording in Rat DRG Neurons.
L4 and L5 DRG neurons were enzymatically dissociated (57), plated onto polylysine-coated glass coverslips, and maintained overnight at 37 °C in a 5% (vol/vol) CO2 incubator in minimum essential media with 10% (vol/vol) FBS and 1% penicillin/streptomycin. DRG neurons were recorded in a solution containing 145 mM N-methyl-d-glucosamine (NMG)⋅Cl, 5 mM BaCl2, 10 mM Na⋅Hepes, and 5 mM glucose, with pH 7.4 and an osmolarity of 320 mOsm. The pipette solution contained 104 mM NMG⋅Cl, 14 mM creatine⋅PO4, 6 mM MgCl2, 10 mM NMG⋅Hepes, 5 mM Tris⋅ATP, 10 mM NMG2⋅EGTA, and 0.3 Tris2⋅GTP, with pH 7.4 and an osmolarity of 300 mOsm. Ionic currents were recorded and analyzed as previously described (19, 58). Recordings were carried out at room temperature, and the holding potential was −80 mV. Group data were calculated as mean ± SD, and Student’s t test was used to determine significant differences (P < 0.05).
NMR Spectroscopy.
Samples were dissolved in 90% (vol/vol) H2O/10% (vol/vol) D2O at pH ∼3, and spectra (total correlation spectroscopy, NOESY, double quantum-filtered correlation spectroscopy, heteronuclear single-quantum correlation spectroscopy, and diffusion-ordered spectroscopy) were recorded on a Bruker 600- or 900-MHz spectrometer at 280 K, 298 K, or 308 K. The samples were also reexamined after addition of 10–30% (vol/vol) TFE. Spectra were acquired with 4,096 data points in the first dimension and 512 data points in the second dimension. Chemical shifts were referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonate at 0 ppm. Spectra were processed with Topspin (Bruker Biospin) and analyzed with SPARKY software. Diffusion experiments were performed to measure the translational diffusion coefficients relative to an internal standard of dioxane (59). The effective hydrodynamic radii of the isomers were measured to determine whether the peptide was aggregating under the conditions used to acquire the NMR data.
Structure calculations were based on distance restraints derived from NOESY spectra, and on backbone dihedral angle restraints generated using TALOS+ (60). A family of structures consistent with the experimental restraints was calculated using CYANA (61). Structures were visualized using MOLMOL (62) and assessed using PROCHECK-NMR (63) and PROMOTIF (64).
CCI.
Male Sprague–Dawley rats (∼200 g) were obtained from the Guangdong Medical Experiment Animal Center and used with permit SCXK 2013-0002 (Guangdong rats) or from the Beijing Si Bei Fu Experiment Animal Company and used with permit SCXK 2011-0004 (Beijing rats). All procedures were reviewed and approved by the Institutional Animal Care and Use Committee of Hainan University and A. T. Still University and followed NIH guidelines. CCI was produced using the method of Bennett and Xie (25). Adult rats were deeply anesthetized with sodium pentobarbital (80 mg/kg i.p.); the right-side sciatic nerve of each animal was exposed at midthigh level, and four chromic gut ligatures (4-0) were tied loosely around the nerve, 1 mm apart. Great care was taken to tie the ligations to constrict the nerve just barely to prevent arresting of the epineural blood flow. The muscle layer and skin were sutured and closed separately. Testing was begun 11–13 d after surgery. By this time, the CCI rats had grown to a weight of ∼250 g.
Mechanical Hyperalgesia Testing.
Mechanical PWT was measured by two methods. One was using a series of manual von Frey hairs (Aesthesio Precision Tactile Sensory Evaluator; Danmic Global) for Beijing rats (65). Another was using an electronic von Frey aesthesiometer (electronic von Frey model 2390 series; IITC Life Sciences) for Guangdong rats. Mechanical PWT (grams) was recorded from the injured right-hind paw of each CCI rat. The CCI rats were randomly divided into different groups to receive i.m. injection of compound. Negative control animals received injections of saline. GeXIVA[1,2] was dissolved in 0.9% saline on the day of the experiment. Four groups of Beijing rats (n = 8) were injected with either saline alone or one of three doses of the peptide GeXIVA[1,2] (0.3 nmol, 0.6 nmol, or 1.2 nmol), and mechanical PWTs were tested using a series of manual von Frey hairs. Another four groups of Guangdong rats (n = 6) were injected with saline alone, 500 nmol of morphine, or GeXIVA[1,2] (1 nmol or 2 nmol). The experimenters were blinded to drug treatments in all experiments.
Molecular Modeling.
The NMR structure of GeXIVA[1,4] was docked into the α9α10 orthosteric binding site of the rat α9α10 nAChR using Modeler 9v13 (66) and structural alignment of its α-helical segment to the α-helical segment of α-conotoxin PnIA bound to AChBP (Protein Data Bank ID code 2BR8), which is a structural surrogate to nAChRs. Two models corresponding to opposite orientations of the α-helix were generated and refined using 20-ns molecular dynamics simulations carried out with GROMACS 4.6.5 (67) and the GROMOS 54a7 force-field (68). The system was heated from 60 to 300 K, and restraints on the positions of the protein atoms were progressively removed over 5 ns. Unconstrained coarse-grained simulations of the interactions of GeXIVA with the ligand-binding domain of α9α10 nAChR were carried out by initially placing GeXIVA randomly around the ligand-binding domain. These simulations were done using GROMACS 4.6.5 (67) and the MARTINI 2.2 force-field (69).
Measurement of Motor Function.
The accelerating rotarod (model 3375-R4; TSE Systems) was used to test αO-conotoxin GeXIVA[1,2] effects on motor function with a maximal cutoff time of 300 s by methods described by Klimis et al. (70). Ambulation was tested by measuring the latency to fail negotiation of accelerating speeds of 4–40 rpm during a maximum time of 300 s. Each animal was tested immediately before i.m. injection, and then at 1, 2, 4, and 6 h after injection. The rats were tested for 300-s trials with an interval of 15 min.
Data Analysis of CCI and Rotarod Tests.
Results of CCI and rotarod tests were expressed as mean ± SEM, which were analyzed using GraphPad Prism (version 5 for Windows). Two-way ANOVA (time and drug) was performed with Bonferroni posttests for CCI PWTs and the latencies of the rotarod test. AUC data were generated using the AUC function in Prism, which were analyzed by one-way ANOVA with Dunnett’s multiple comparison posttests, including the preinjection (t = 0) to 6-h time point. P < 0.05 was considered statistically significant.
SI Materials and Methods
Materials.
RP-HPLC analytical Vydac C18 column (5 μm, 4.6 mm × 250 mm) and preparative C18 Vydac column (10 μm, 22 mm × 250 mm) were from Grace Vydac. Reagents for peptide synthesis were from GL Biochem. Clones of rat α2–α7 and β4, as well as mouse muscle α1, β1, δ-, and ε-subunit cDNAs were kindly provided by S. Heinemann (Salk Institute, San Diego, CA). Clones for α9 and α10 were generously provided by A. B. Elgoyhen (Buenos Aires, Argentina). Clones of β2 and β3 subunits in the high-expressing pGEMHE vector were kindly provided by C. W. Luetje (University of Miami, Miami, FL).
cDNA Cloning and Sequencing.
RT-PCR conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, and a final step at 72 °C for 2 min. The resulting PCR fragments were cloned and sequenced.
Peptide Synthesis.
Peptides were cleaved from the solid support by treatment with reagent K [TFA∕water∕ethanedithiol∕phenol∕thioanisole: 81.82/4.55/2.27/6.82/4.55 (vol∕vol∕vol∕vol∕vol)]. Released peptides were precipitated and washed three times with cold ether. The monocyclic and bicyclic peptides were purified by HPLC on an RP C18 Vydac column using a linear gradient of 10–50% B60 for 40 min. Solvent B was 60% ACN, 40% H20, and 0.092% TFA; solvent A was 0.1% TFA in H2O. Peptide concentration was measured using a spectrophotometer at a wavelength of 280 nM and calculated using the Beer–Lambert equation.
Voltage-Clamp Recording.
The oocyte chamber consisting of a cylindrical well (∼30 μL in volume) was gravity-perfused at a rate of ∼2 mL⋅min−1 with ND96 buffer containing 0.1 mg/mL BSA. ND96 buffer consisted of 96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, and 5 mM Hepes (pH 7.1–7.5). The Ba++ ND96 had 1.8 mM BaCl2 in place of CaCl2. The oocyte was subjected once a minute to a 1-s pulse of ACh. The 100 μM ACh pulse was used for most subtypes, except for α9α10 (10 μM), muscle α1β1δε (10 μM), and α7 (200 μM) subtypes. For toxin concentrations of 10 μM and lower, once a stable baseline was achieved, either ND-96 alone or ND-96 containing conotoxin was manually preapplied for 5 min before addition of the agonist.
Data Analysis.
The average of five control responses just preceding a test response was used to normalize the test response to obtain “% response.” Each data point of a dose–response curve represents the average ± SE of at least three oocytes. The dose–response data were fit to an equation, % response = 100/[1 + ([toxin]/IC50) ^ nH], where nH is the Hill coefficient, by nonlinear regression analysis using GraphPad Prism. Responses to ACh of α9α10 nAChR in ND96 or Ba++ ND96 were calculated as the percentage of the responses observed at −90 mV when changing holding membrane potentials from −90 mV to +30 mV by 10-mV steps once every 1 min. The voltage current curves were plotted with Prism software.
Voltage-Dependent Calcium Current Recording in Rat DRG Neurons.
All procedures were reviewed and approved by the Institutional Animal Care and Use Committee and followed NIH guidelines. Rats were killed with CO2, followed by decapitation. Solutions were applied using a gravity-fed perfusion system with six inputs and a single output. The solution exchange time was 2 s for this system. Cell diameter was calculated using the cell capacitance and assuming a specific membrane capacitance of 1 μF/μm2.
CCI.
Rats were housed and maintained with a standard 12-h light/dark cycle and free access to food and water.
Mechanical Hyperalgesia Testing.
Rats were placed in elevated organic glass cages with 3-mm × 3-mm wire mesh bases suspended above a table. All rats were given 30 min to acclimate to the testing environment and to become relaxed and stationary. Hyperalgesia testing was performed several times before CCI (baseline) and at different time points after CCI.
Measurement of Motor Function.
All of the rats were trained for 2 d, twice a day, 30 min per time before testing. Trained eligible rats were divided into two groups (n = 8; i.e., drug αO-conotoxin GeXIVA[1,2] and vehicle saline). Latency of each rat to fall off from the accelerating rotarod was determined. Each latency recording consisted of the mean of three measurements.
Acknowledgments
We thank Baldomero Olivera, Layla Azam, Doju Yoshikami, Ruibin Su, and Olivier Cheneval for advice and help. This work was supported, in part, by the Major International Joint Research Project of the National Natural Science Foundation of China (Grant 81420108028), the Program for International Science and Technology Cooperation Program of China (Grant 2011DFR31210), the State High-Tech Research and Development Project (863) of the Ministry of Science and Technology of China (Grant 2012AA021706), the National Natural Science Foundation of China (Grants 81160503 and 41366002), and Changjiang Scholars and Innovative Research Team in University Grant IRT1123. This work was also supported by NIH Grants GM103801 and GM48677 and by Australian Research Council Grant 1093115. D.J.C. is a National Health and Medical Research Council Professorial Fellow (Grants APP1026501 and APP1076136). We also acknowledge financial support provided by the Queensland State Government to the Queensland NMR Network facilities at The University of Queensland, Australia. V.I.T. was supported by Russian Scientific Foundation Grant 14-24-00118.
Footnotes
Conflict of interest statement: The sequence of αO-conotoxin GeXIVA has been patented by Hainan University, with S.L., D.Z., Y.W., X.Z., Y.H., and J.M.M. listed as inventors.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KP793740).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503617112/-/DCSupplemental.
References
- 1.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol Interv. 2007;7(5):251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 2.Mayer AM, et al. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol Sci. 2010;31(6):255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Kaas Q, Westermann JC, Craik DJ. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon. 2010;55(8):1491–1509. doi: 10.1016/j.toxicon.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Gotti C, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78(7):703–711. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 5.Beech RN, Callanan MK, Rao VT, Dawe GB, Forrester SG. Characterization of cys-loop receptor genes involved in inhibitory amine neurotransmission in parasitic and free living nematodes. Parasitol Int. 2013;62(6):599–605. doi: 10.1016/j.parint.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Elgoyhen AB, Katz E. The efferent medial olivocochlear-hair cell synapse. J Physiol Paris. 2012;106(1-2):47–56. doi: 10.1016/j.jphysparis.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colomer C, et al. Functional characterization of alpha9-containing cholinergic nicotinic receptors in the rat adrenal medulla: Implication in stress-induced functional plasticity. J Neurosci. 2010;30(19):6732–6742. doi: 10.1523/JNEUROSCI.4997-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simard AR, et al. Differential modulation of EAE by α9*- and β2*-nicotinic acetylcholine receptors. Immunol Cell Biol. 2013;91(3):195–200. doi: 10.1038/icb.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIntosh JM, Absalom N, Chebib M, Elgoyhen AB, Vincler M. Alpha9 nicotinic acetylcholine receptors and the treatment of pain. Biochem Pharmacol. 2009;78(7):693–702. doi: 10.1016/j.bcp.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipovsek M, et al. Phylogenetic differences in calcium permeability of the auditory hair cell cholinergic nicotinic receptor. Proc Natl Acad Sci USA. 2012;109(11):4308–4313. doi: 10.1073/pnas.1115488109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franchini LF, Elgoyhen AB. Adaptive evolution in mammalian proteins involved in cochlear outer hair cell electromotility. Mol Phylogenet Evol. 2006;41(3):622–635. doi: 10.1016/j.ympev.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 12.Horne WA, Hawrot E, Tsien RW. omega-Conotoxin GVIA receptors of Discopyge electric organ. Characterization of omega-conotoxin binding to the nicotinic acetylcholine receptor. J Biol Chem. 1991;266(21):13719–13725. [PubMed] [Google Scholar]
- 13.McIntosh JM, et al. A new family of conotoxins that blocks voltage-gated sodium channels. J Biol Chem. 1995;270(28):16796–16802. doi: 10.1074/jbc.270.28.16796. [DOI] [PubMed] [Google Scholar]
- 14.Katz E, et al. High calcium permeability and calcium block of the alpha9 nicotinic acetylcholine receptor. Hear Res. 2000;141(1-2):117–128. doi: 10.1016/s0378-5955(99)00214-2. [DOI] [PubMed] [Google Scholar]
- 15.Elgoyhen AB, et al. alpha10: A determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc Natl Acad Sci USA. 2001;98(6):3501–3506. doi: 10.1073/pnas.051622798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azam L, McIntosh JM. Molecular basis for the differential sensitivity of rat and human α9α10 nAChRs to α-conotoxin RgIA. J Neurochem. 2012;122(6):1137–1144. doi: 10.1111/j.1471-4159.2012.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellison M, et al. Alpha-RgIA, a novel conotoxin that blocks the alpha9alpha10 nAChR: Structure and identification of key receptor-binding residues. J Mol Biol. 2008;377(4):1216–1227. doi: 10.1016/j.jmb.2008.01.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CS, Song JH, Nagata K, Yeh JZ, Narahashi T. Effects of the neuroprotective agent riluzole on the high voltage-activated calcium channels of rat dorsal root ganglion neurons. J Pharmacol Exp Ther. 1997;282(3):1280–1290. [PubMed] [Google Scholar]
- 19.Ramachandra R, et al. Identification of CaV channel types expressed in muscle afferent neurons. J Neurophysiol. 2013;110(7):1535–1543. doi: 10.1152/jn.00069.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu SG, Zhang XL, Luo ZD, Gold MS. Persistent inflammation alters the density and distribution of voltage-activated calcium channels in subpopulations of rat cutaneous DRG neurons. Pain. 2010;151(3):633–643. doi: 10.1016/j.pain.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5(1):67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 22.Akondi KB, et al. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem Rev. 2014;114(11):5815–5847. doi: 10.1021/cr400401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pucci L, et al. Engineering of α-conotoxin MII-derived peptides with increased selectivity for native α6β2* nicotinic acetylcholine receptors. FASEB J. 2011;25(11):3775–3789. doi: 10.1096/fj.10-179853. [DOI] [PubMed] [Google Scholar]
- 24.Austin PJ, Wu A, Moalem-Taylor G. Chronic constriction of the sciatic nerve and pain hypersensitivity testing in rats. J Vis Exp. 2012;(61):3393. doi: 10.3791/3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 26.Sanford M. Intrathecal ziconotide: A review of its use in patients with chronic pain refractory to other systemic or intrathecal analgesics. CNS Drugs. 2013;27(11):989–1002. doi: 10.1007/s40263-013-0107-5. [DOI] [PubMed] [Google Scholar]
- 27.Di Cesare Mannelli L, et al. Α-conotoxin RgIA protects against the development of nerve injury-induced chronic pain and prevents both neuronal and glial derangement. Pain. 2014;155(10):1986–1995. doi: 10.1016/j.pain.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams DJ, Callaghan B, Berecki G. Analgesic conotoxins: Block and G protein-coupled receptor modulation of N-type (Ca(V) 2.2) calcium channels. Br J Pharmacol. 2012;166(2):486–500. doi: 10.1111/j.1476-5381.2011.01781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callaghan B, et al. Analgesic alpha-conotoxins Vc1.1 and Rg1A inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J Neurosci. 2008;28(43):10943–10951. doi: 10.1523/JNEUROSCI.3594-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuny H, et al. γ-Aminobutyric acid type B (GABAB) receptor expression is needed for inhibition of N-type (Cav2.2) calcium channels by analgesic α-conotoxins. J Biol Chem. 2012;287(28):23948–23957. doi: 10.1074/jbc.M112.342998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daly NL, et al. Structure and activity of alpha-conotoxin PeIA at nicotinic acetylcholine receptor subtypes and GABA(B) receptor-coupled N-type calcium channels. J Biol Chem. 2011;286(12):10233–10237. doi: 10.1074/jbc.M110.196170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh TG, Cuny H, Slesinger PA, Adams DJ. Novel mechanism of voltage-gated N-type (Cav2.2) calcium channel inhibition revealed through alpha-conotoxin Vc1.1 activation of the GABA(B) receptor. Mol Pharmacol. 2015;87(2):240–250. doi: 10.1124/mol.114.096156. [DOI] [PubMed] [Google Scholar]
- 33.van Lierop BJ, et al. Dicarba α-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors. ACS Chem Biol. 2013;8(8):1815–1821. doi: 10.1021/cb4002393. [DOI] [PubMed] [Google Scholar]
- 34.Wright ABNY, McIntosh JM, Elmslie KS. Limited efficacy of α-conopeptides, Vc1.1 and RgIA, to inhibit sensory neuron CaV current. eNeuro. 2015 doi: 10.1523/ENEURO.0057-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammadi S, Christie MJ. α9-nicotinic acetylcholine receptors contribute to the maintenance of chronic mechanical hyperalgesia, but not thermal or mechanical allodynia. Mol Pain. 2014;10:64. doi: 10.1186/1744-8069-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azam L, et al. Molecular interaction of α-conotoxin RgIA with the rat α9α10 nicotinic acetylcholine receptor. Mol Pharmacol. 2015;87(5):855–864. doi: 10.1124/mol.114.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutton JL, et al. A new level of conotoxin diversity, a non-native disulfide bond connectivity in alpha-conotoxin AuIB reduces structural definition but increases biological activity. J Biol Chem. 2002;277(50):48849–48857. doi: 10.1074/jbc.M208842200. [DOI] [PubMed] [Google Scholar]
- 38.Gao B, et al. Expression, renaturation and biological activity of recombinant conotoxin GeXIVAWT. Appl Microbiol Biotechnol. 2013;97(3):1223–1230. doi: 10.1007/s00253-012-4287-6. [DOI] [PubMed] [Google Scholar]
- 39.Luo S, et al. Diversity of the O-superfamily conotoxins from Conus miles. J Pept Sci. 2007;13(1):44–53. doi: 10.1002/psc.802. [DOI] [PubMed] [Google Scholar]
- 40.Möller C, et al. A novel conotoxin framework with a helix-loop-helix (Cs alpha/alpha) fold. Biochemistry. 2005;44(49):15986–15996. doi: 10.1021/bi0511181. [DOI] [PubMed] [Google Scholar]
- 41.Nicke A. Learning about structure and function of neuronal nicotinic acetylcholine receptors. Lessons from snails. Eur J Biochem. 2004;271(12):2293. doi: 10.1111/j.1432-1033.2004.04171.x. [DOI] [PubMed] [Google Scholar]
- 42.Davis J, Jones A, Lewis RJ. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides. 2009;30(7):1222–1227. doi: 10.1016/j.peptides.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Vincler M, et al. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc Natl Acad Sci USA. 2006;103(47):17880–17884. doi: 10.1073/pnas.0608715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satkunanathan N, et al. Alpha-conotoxin Vc1.1 alleviates neuropathic pain and accelerates functional recovery of injured neurones. Brain Res. 2005;1059(2):149–158. doi: 10.1016/j.brainres.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Del Bufalo A, Cesario A, Salinaro G, Fini M, Russo P. Alpha9 alpha10 nicotinic acetylcholine receptors as target for the treatment of chronic pain. Curr Pharm Des. 2014;20(38):6042–6047. doi: 10.2174/1381612820666140314150634. [DOI] [PubMed] [Google Scholar]
- 46.McIntosh JM, et al. A novel alpha-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat alpha9alpha10 and alpha7 nicotinic cholinergic receptors. J Biol Chem. 2005;280(34):30107–30112. doi: 10.1074/jbc.M504102200. [DOI] [PubMed] [Google Scholar]
- 47.Holtman JR, et al. The novel small molecule α9α10 nicotinic acetylcholine receptor antagonist ZZ-204G is analgesic. Eur J Pharmacol. 2011;670(2-3):500–508. doi: 10.1016/j.ejphar.2011.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng G, et al. Discovery of non-peptide, small molecule antagonists of α9α10 nicotinic acetylcholine receptors as novel analgesics for the treatment of neuropathic and tonic inflammatory pain. Bioorg Med Chem Lett. 2011;21(8):2476–2479. doi: 10.1016/j.bmcl.2011.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wala EP, Crooks PA, McIntosh JM, Holtman JR., Jr Novel small molecule α9α10 nicotinic receptor antagonist prevents and reverses chemotherapy-evoked neuropathic pain in rats. Anesth Analg. 2012;115(3):713–720. doi: 10.1213/ANE.0b013e31825a3c72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernyavsky AI, Arredondo J, Vetter DE, Grando SA. Central role of alpha9 acetylcholine receptor in coordinating keratinocyte adhesion and motility at the initiation of epithelialization. Exp Cell Res. 2007;313(16):3542–3555. doi: 10.1016/j.yexcr.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chikova A, Grando SA. Naturally occurring variants of human Α9 nicotinic receptor differentially affect bronchial cell proliferation and transformation. PLoS One. 2011;6(11):e27978. doi: 10.1371/journal.pone.0027978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chernyavsky AI, Shchepotin IB, Galitovkiy V, Grando SA. Mechanisms of tumor-promoting activities of nicotine in lung cancer: synergistic effects of cell membrane and mitochondrial nicotinic acetylcholine receptors. BMC Cancer. 2015;15:152. doi: 10.1186/s12885-015-1158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quan Y, Luo S, Lin Q, Zhangsun D, Zhang B. Conotoxin RNA isolation and its cDNA synthesis. Chinese Journal of Marine Drugs. 2005;24(2):1–5. [Google Scholar]
- 54.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17(1):107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 55.Dowell C, et al. Alpha-conotoxin PIA is selective for alpha6 subunit-containing nicotinic acetylcholine receptors. J Neurosci. 2003;23(24):8445–8452. doi: 10.1523/JNEUROSCI.23-24-08445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cartier GE, et al. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271(13):7522–7528. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- 57.Puhl HL, 3rd, Ikeda SR. Identification of the sensory neuron specific regulatory region for the mouse gene encoding the voltage-gated sodium channel NaV1.8. J Neurochem. 2008;106(3):1209–1224. doi: 10.1111/j.1471-4159.2008.05466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yarotskyy V, Gao G, Peterson BZ, Elmslie KS. Domain III regulates N-type (CaV2.2) calcium channel closing kinetics. J Neurophysiol. 2012;107(7):1942–1951. doi: 10.1152/jn.00993.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilkins DK, et al. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38(50):16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 60.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeya T, Terauchi T, Güntert P, Kainosho M. Evaluation of stereo-array isotope labeling (SAIL) patterns for automated structural analysis of proteins with CYANA. Magn Reson Chem. 2006;44(Spec No):S152–S157. doi: 10.1002/mrc.1815. [DOI] [PubMed] [Google Scholar]
- 62.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 63.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 64.Hutchinson EG, Thornton JM. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5(2):212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 66.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 67.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmid N, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J. 2011;40(7):843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 69.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111(27):7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 70.Klimis H, et al. A novel mechanism of inhibition of high-voltage activated calcium channels by α-conotoxins contributes to relief of nerve injury-induced neuropathic pain. Pain. 2011;152(2):259–266. doi: 10.1016/j.pain.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 71.Luo S, et al. A novel inhibitor of α9α10 nicotinic acetylcholine receptors from Conus vexillum delineates a new conotoxin superfamily. PLoS One. 2013;8(1):e54648. doi: 10.1371/journal.pone.0054648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puillandre N, Koua D, Favreau P, Olivera BM, Stöcklin R. Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol. 2012;74(5-6):297–309. doi: 10.1007/s00239-012-9507-2. [DOI] [PubMed] [Google Scholar]
- 73.Dutertre S, et al. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 2013;12(2):312–329. doi: 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nielsen KJ, et al. Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels. J Mol Biol. 1999;289(5):1405–1421. doi: 10.1006/jmbi.1999.2817. [DOI] [PubMed] [Google Scholar]
- 75.Olivera BM, et al. Peptide neurotoxins from fish-hunting cone snails. Science. 1985;230(4732):1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 76.Imperial JS, et al. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45(27):8331–8340. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- 77.Lewis RJ, Dutertre S, Vetter I, Christie MJ. Conus venom peptide pharmacology. Pharmacol Rev. 2012;64(2):259–298. doi: 10.1124/pr.111.005322. [DOI] [PubMed] [Google Scholar]
- 78.Peng C, et al. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27(9):2174–2181. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 79.Lluisma AO, López-Vera E, Bulaj G, Watkins M, Olivera BM. Characterization of a novel psi-conotoxin from Conus parius Reeve. Toxicon. 2008;51(2):174–180. doi: 10.1016/j.toxicon.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shon KJ, et al. A noncompetitive peptide inhibitor of the nicotinic acetylcholine receptor from Conus purpurascens venom. Biochemistry. 1997;36(31):9581–9587. doi: 10.1021/bi970235w. [DOI] [PubMed] [Google Scholar]
- 81.Luo S, et al. Atypical alpha-conotoxin LtIA from Conus litteratus targets a novel microsite of the alpha3beta2 nicotinic receptor. J Biol Chem. 2010;285(16):12355–12366. doi: 10.1074/jbc.M109.079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobsen R, et al. Differential targeting of nicotinic acetylcholine receptors by novel alphaA-conotoxins. J Biol Chem. 1997;272(36):22531–22537. doi: 10.1074/jbc.272.36.22531. [DOI] [PubMed] [Google Scholar]
- 83.Jimenez EC, Olivera BM, Teichert RW. AlphaC-conotoxin PrXA: A new family of nicotinic acetylcholine receptor antagonists. Biochemistry. 2007;46(30):8717–8724. doi: 10.1021/bi700582m. [DOI] [PubMed] [Google Scholar]
- 84.Loughnan M, et al. Identification of a novel class of nicotinic receptor antagonists: Dimeric conotoxins VxXIIA, VxXIIB, and VxXIIC from Conus vexillum. J Biol Chem. 2006;281(34):24745–24755. doi: 10.1074/jbc.M603703200. [DOI] [PubMed] [Google Scholar]
- 85.Teichert RW, Jimenez EC, Olivera BM. Alpha S-conotoxin RVIIIA: A structurally unique conotoxin that broadly targets nicotinic acetylcholine receptors. Biochemistry. 2005;44(21):7897–7902. doi: 10.1021/bi047274+. [DOI] [PubMed] [Google Scholar]
- 86.Biggs JS, et al. Evolution of Conus peptide toxins: Analysis of Conus californicus Reeve, 1844. Mol Phylogenet Evol. 2010;56(1):1–12. doi: 10.1016/j.ympev.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhangsun D, et al. Novel O-superfamily conotoxins identified by cDNA cloning from three vermivorous Conus species. Chem Biol Drug Des. 2006;68(5):256–265. doi: 10.1111/j.1747-0285.2006.00443.x. [DOI] [PubMed] [Google Scholar]
- 88.Azam L, et al. Alpha-conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280(1):80–87. doi: 10.1074/jbc.M406281200. [DOI] [PubMed] [Google Scholar]
- 89.Hopkins C, et al. A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J Biol Chem. 1995;270(38):22361–22367. doi: 10.1074/jbc.270.38.22361. [DOI] [PubMed] [Google Scholar]
- 90.Loughnan ML, Nicke A, Lawrence N, Lewis RJ. Novel alpha D-conopeptides and their precursors identified by cDNA cloning define the D-conotoxin superfamily. Biochemistry. 2009;48(17):3717–3729. doi: 10.1021/bi9000326. [DOI] [PubMed] [Google Scholar]
- 91.Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24(3):445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]