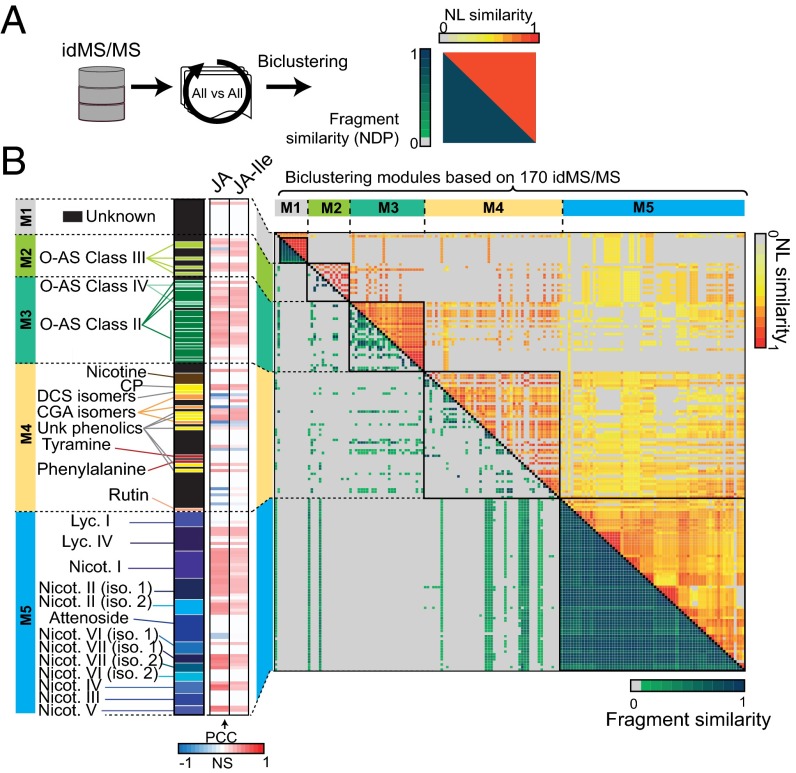
Fig. 4.
Biclustering of idMS/MS according to structural relationships computed from fragment and neutral loss similarity metrics facilitate compound class assignments. (A) All-against-all alignment of idMS/MS based on fragment and neutral loss similarity calculation. (B) Biclustering using the R Diffcoex package of idMS/MS according to results of these two similarity analyses identifies five idMS/MS modules (M1, M2, M3, M4, and M5) that partly overlap to a priori knowledge on compound class definition. Green-to-blue gradient denotes medium-to-high fragment similarity whereas that from yellow-to-red indicates medium-to-high neutral loss similarity. Compound annotation is indicated on the left of the heatmap. Black cells correspond to unknown metabolites whereas the different color variations correspond to different compound classes. The next heatmap bar visualizes significant Pearson correlation values with JA and JA-Ile induced levels as detected in Fig. 3. A neutral loss (NL) map in which shared NLs between classified idMS/MS are reported is presented in SI Appendix, Fig. S6. This map was used to infer NLs overrepresented in a particular module. Close views on module subsections highlighting shared NLs and relevant m/z features resulting from fragmentation are reported in SI Appendix, Figs. S7 and S8. CGA, chlorogenic acid; DCS, N′,N′′-dicaffeoylspermidine; iso., isomer; Lyc., lyciumoside; Nicot., nicotianoside; O-AS, O-acyl sugars.