Fig. 5.
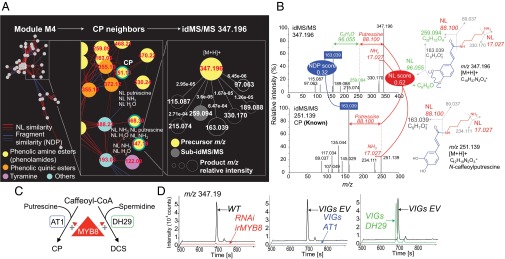
Navigating the idMS/MS similarity network supports structural predictions for novel herbivory-regulated metabolites. (A) Close-up views on the similarity network constructed for module 4 resulting from the biclustering analysis. Module 4 is enriched in previously characterized and structurally elucidated phenolamides, most of which are strongly responsive to simulated herbivory treatments, but also includes unknown metabolites with JA signaling associated natural variation such as m/z 347.19 at retention time 245 s. The composite idMS/MS for m/z 347.19 is the one of the first network neighbors of N-caffeoylputrescine (CP) with idMS/MS m/z 251.13 due to neutral loss and fragment-based similarities. (B) idMS/MS m/z 251.13 and m/z 347.19 share neutral loss corresponding to the loss of putrescine. The intense fragment peak at m/z 163.04 shared by both idMS/MS corresponds to the caffeoyl moiety cleavage from the putrescine. Interestingly, in the case of idMS/MS m/z 347.19, only this fragment derives from an additional neutral loss of C6H8O as part of pseudo-MS3 reaction supported by the alignment of idMS/MS m/z 259.13. (C) Working model for MYB8-regulated N-acyltransferase–mediated production of phenolamides. AT1 catalyzes the formation of putrescine-based phenolamides whereas DH29 acts as a first committed step in spermidine conjugate production. (D) Extracted ion traces for m/z 347.19 supporting its classification as an MYB8-dependent, putrescine-based phenolamide dependent on the catalytic activity of AT1 (SI Appendix, Table S3). Additional results obtained from molecular studies of the metabolic conversion from CP to the phenolamide-related m/z 347.19 are presented in SI Appendix, Fig. S10. EV, empty-vector VIGs control; irMYB8, stably silenced MYB8 transformant; VIGs, virus-induced gene transient silencing.
