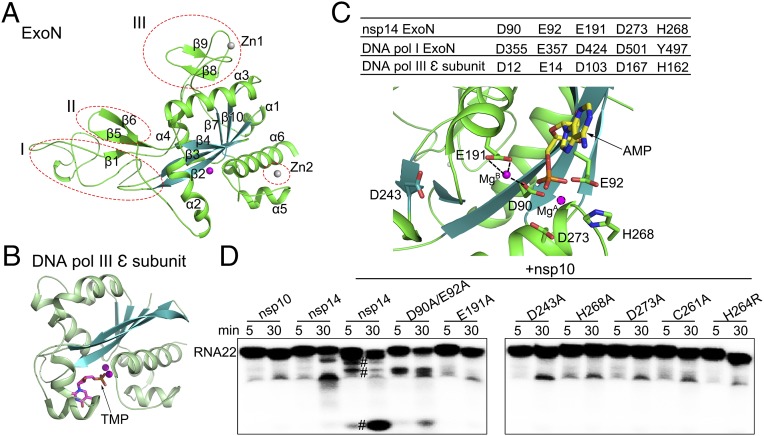
Fig. 2.
Comparison of the structure and catalytic residues of nsp14 ExoN domain with proofreading homologs. (A) Cartoon representation of the ExoN domain marked with secondary structural elements. The three different regions from other DEDD superfamily exonucleases are indicated by red dashed ellipses. (B) The structure of the E. coli ε subunit of polymerase III (Pol III) is shown in the same orientation as nsp14 for comparison. Metal ions are shown as spheres, and bound ligands are shown as sticks. (C) The active center of the ExoN domain of nsp14. (Upper) Catalytic residues of the ExoN domain of nsp14, the exonuclease domain of DNA polymerase I, and the ε subunit of DNA polymerase III of E. coli are listed in the table. (Lower) Catalytic residues, the modeled substrate AMP, and the mistaken D243 are shown as sticks. MgB observed in the structure and MgA modeled are shown as spheres. Dashed lines indicate the hydrogen bonds between MgB and D90 and E191. (D) Exoribonuclease assays for nsp10, nsp14 alone, and nsp14 or nsp14 mutants in complex with nsp10 on 5′-labeled ssRNA of 22 nucleosides (RNA22). The symbol “#” indicates cleavage products.