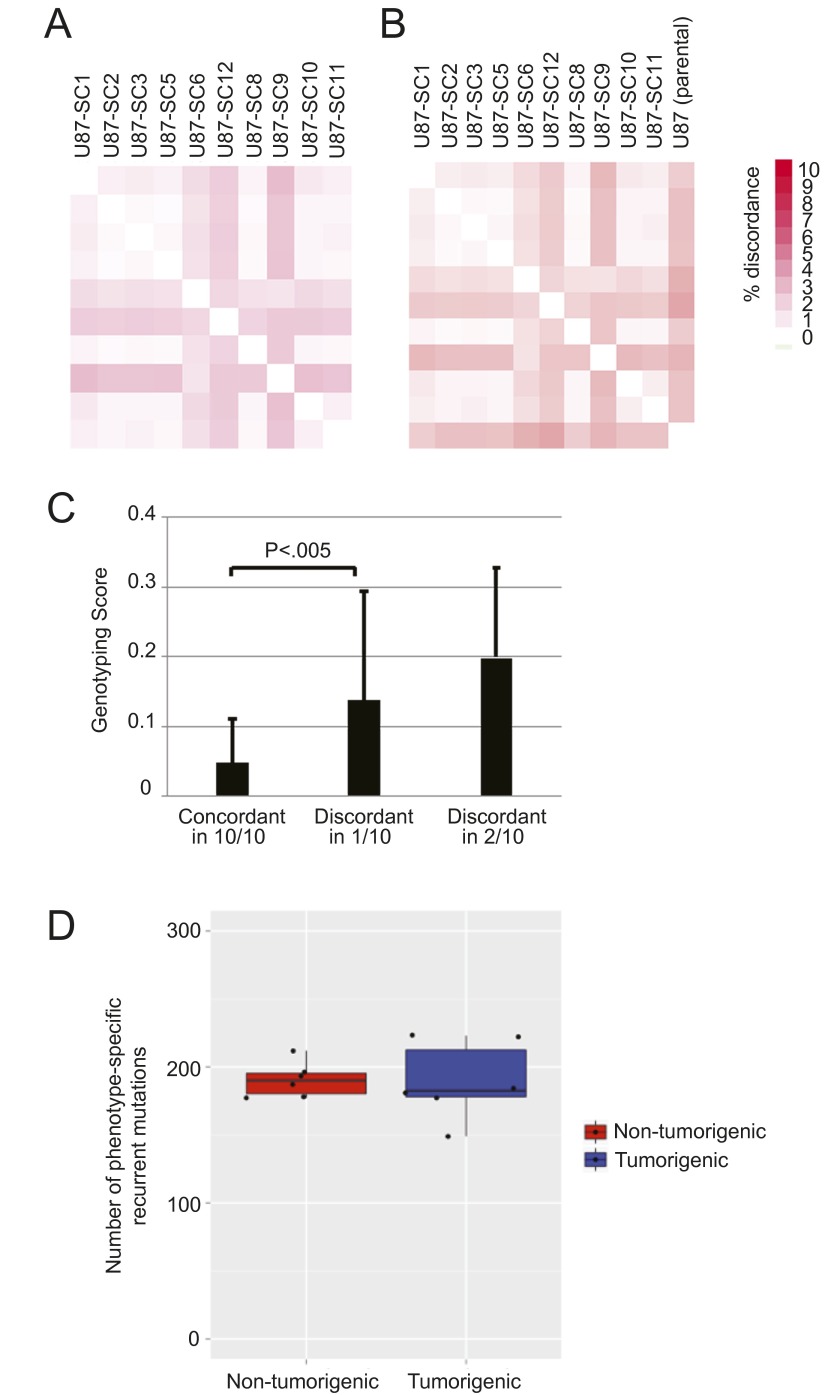
Fig. S1.
Differences in glioblastoma tumorigenicity among U87MG subclones are unlikely due to genetic changes. (A) Genotyping of U87MG subclones using Affymetrix Human Mapping 250K NSP arrays. Heat map representing the fraction of discordant genotypes between each pair of clones out of ∼251,650 loci called in both samples. (B) Heat map representing the fraction of discordant genotypes between each clone and the parental U87MG cell line at the ∼80,398 loci called in both Illumina and Affymetrix arrays. (C) Genotype quality score (decreasing scale) between scores at 228,774 × 10 genotypes concordant in all 10 clones and scores at 2,435 and 920 × 2 genotypes discordant in one or two clones, respectively. Statistical differences between groups were assessed using the paired Student’s t test; error bars represent SD. Of all the loci, 98% were concordant in all tested U87MG subclones; the low level of discordance is attributable to systematic errors in the genotyping, as the quality scores of the concordant genotypes were significantly different from those of discordant genotypes. (D) Number of recurrent mutations specific to nontumorigenic versus tumorigenic subclones of the 83 and U87MG glioblastoma cell lines, determined by whole exome sequencing of six subclones of each phenotype. P = 0.55 by Student’s t test, showing no significant difference that would otherwise suggest tumorigenesis is linked to a gross difference in mutational load.