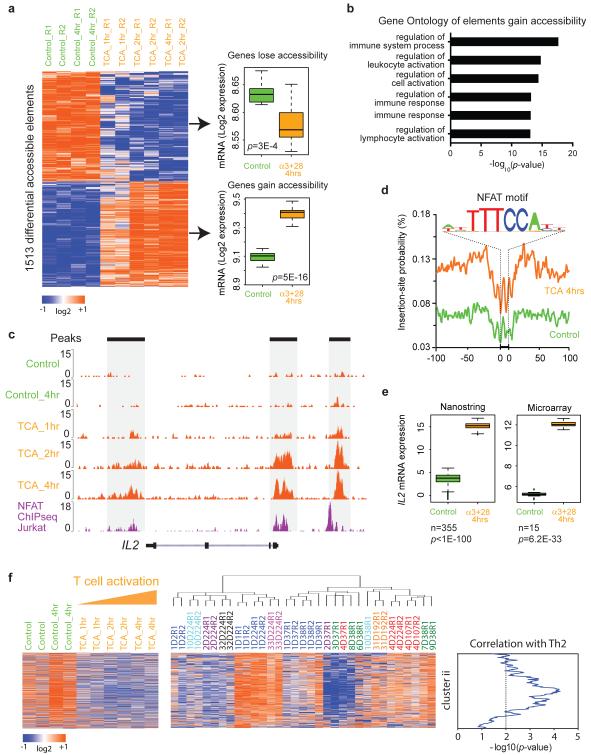
Figure 5. Regulome dynamics during T cell activation.
a, Left: heat map showing time course of regulatory elements with differential ATAC-seq activity during T cell activation. Each column is a sample; each row is an element. Samples and elements are organized by supervised hierarchical clustering. Color scale indicates relative ATAC-seq signal as indicated. Samples from untreated control and 4 hours control are colored green, and T cell activation at 1, 2, and 4 hours are colored orange. Right: box-plots of mRNA expression levels of the indicated genes in untreated control CD4+ T cells or after 4 hours activation with anti-CD3/CD28. Number of replicates = 15. P-value estimated from Student t-test.
b, Gene Ontology terms of regulatory elements gain in accessibility during T cell activation.
c, Dynamics of ATAC-seq signal at IL2 locus (orange track) during T cell activation (TCA) at the indicated times. NFAT ChIP-seq data in Jurkat cells (purple track) is shown for comparison.
d, Visualization of ATAC-seq footprint for transcription factor NFAT (motif shown) in control cells (green) vs. cells after T cell activation for 4 hours (orange). ATAC-seq signal across all NFAT binding sites in the genome were aligned on the motif and averaged.
e, Box-plots of mRNA expression levels of IL2 in untreated control (green) and 4 hours T cell activation with anti-CD3/CD28 (orange), obtained from Nanostring (left) and microarray (right). mRNA level from 355 or 15 healthy donors were measured by Nanostring or microarray, respectively. Log2 data are shown; P-value estimated from Student t-test.
f, Use of regulome signature of T cell activation to interpret individual variation. Regulatory element accessibility during T cell activation in vitro (left) vs. personal variation from healthy donors (right) are shown. Cluster ii from Figure 1b is found to exhibit coordinate deactivation during T cell activation and is also correlated with a Th2 signature. Donor sample dendrogram and demographic correlation are as in Figure 1b.