Abstract
Aims: Recent studies suggest that the molybdenum enzymes xanthine oxidase, aldehyde oxidase, and mARC exhibit nitrite reductase activity at low oxygen pressures. However, inhibition studies of xanthine oxidase in humans have failed to block nitrite-dependent changes in blood flow, leading to continued exploration for other candidate nitrite reductases. Another physiologically important molybdenum enzyme—sulfite oxidase (SO)—has not been extensively studied. Results: Using gas-phase nitric oxide (NO) detection and physiological concentrations of nitrite, SO functions as nitrite reductase in the presence of a one-electron donor, exhibiting redox coupling of substrate oxidation and nitrite reduction to form NO. With sulfite, the physiological substrate, SO only facilitates one turnover of nitrite reduction. Studies with recombinant heme and molybdenum domains of SO indicate that nitrite reduction occurs at the molybdenum center via coupled oxidation of Mo(IV) to Mo(V). Reaction rates of nitrite to NO decreased in the presence of a functional heme domain, mediated by steric and redox effects of this domain. Using knockdown of all molybdopterin enzymes and SO in fibroblasts isolated from patients with genetic deficiencies of molybdenum cofactor and SO, respectively, SO was found to significantly contribute to hypoxic nitrite signaling as demonstrated by activation of the canonical NO-sGC-cGMP pathway. Innovation: Nitrite binds to and is reduced at the molybdenum site of mammalian SO, which may be allosterically regulated by heme and molybdenum domain interactions, and contributes to the mammalian nitrate-nitrite-NO signaling pathway in human fibroblasts. Conclusion: SO is a putative mammalian nitrite reductase, catalyzing nitrite reduction at the Mo(IV) center. Antioxid. Redox Signal. 23, 283–294.
Introduction
While nitrate and nitrite were traditionally considered inert and potentially toxic metabolites of nitric oxide (NO) oxidation, it is now increasingly appreciated that both nitrate and nitrite can be recycled to bioactive NO (10, 12, 28). Studies have suggested that this reductive mammalian nitrate-nitrite-NO pathway mediates important signaling events and, as such, complements the traditional oxidative L-arginine-NO synthase-NO pathway (29). Nitrate is formed in the blood from oxidation of NO by oxyhemoglobin or is derived from dietary sources, such as leafy green vegetables and water. While the majority of absorbed inorganic nitrate is ultimately excreted into the urine, approximately 25% of plasma nitrate is taken up by the salivary glands and concentrated 10- to 20- fold in saliva, providing a route for entero-salivary recycling of nitrate (30, 34, 52). Symbiotic bacteria in the oral cavity effectively reduce nitrate to nitrite, which is further reduced to NO in the acidic stomach or systemically absorbed (30, 34, 50). Approximately half of plasma nitrite is derived from reduction of dietary nitrate by oral bacteria, whereas half is derived from the oxidation of NO synthase-generated NO in plasma by ceruloplasmin (36, 39). Nitrite is bioactive; it is reduced to NO to regulate blood pressure (10, 12, 13, 50), hypoxic vasodilation (10, 16, 31), cellular cytoprotection (34), and mitochondrial respiration; and functions under hypoxic or exercise stress (24, 40, 51). A central current focus in this field aims at elucidating the processes and enzymes contributory to the one-electron and one-proton transfer reaction of nitrite to form NO (Eq. 1).
 |
Innovation.
Nitrite binds to and is reduced at the molybdenum site of mammalian sulfite oxidase (SO) to form nitric oxide (NO), and, thus, contributes to the mammalian nitrate-nitrite-NO signaling pathway.
Such proton and electron transfer reactions can be catalyzed by redox-active hemoproteins, such as hemoglobin, myoglobin, neuroglobin, cytoglobin, and plant hemoglobins (10, 19, 40, 44, 45, 47), and can also be catalyzed by molybdenum cofactor (Moco−)-containing enzymes, such as xanthine oxidase, aldehyde oxidase, and mARC (2, 26, 41). However, despite evidence that these Moco enzymes can contribute to nitrite-dependent signaling in rodents, studies in humans have shown that infusions of allopurinol or oxypurinol, specific xanthine oxidoreductase inhibitors, do not sufficiently block nitrite-mediated vasodilation (12). Therefore, we evaluated sulfite oxidase (SO), a third Moco-containing enzyme, as a potential nitrite reductase and further examined the catalytic mechanism in the purified enzyme system.
In contrast to xanthine oxidase and aldehyde oxidase, eukaryotic nitrate reductase in plants and SO in animals and plants represent a distinct second family of Moco enzymes that catalyze oxygen atom transfer reactions (43). For plant nitrate reductase, nitrite-dependent NO synthesis has been reported (53). Given the high structural similarity between nitrate reductase and SO (9), it is likely that nitrite would react with the reduced molybdenum center of SO. SO is a homodimer (110 kDa) with each monomer containing an N-terminal cytochrome b5-type heme, a central Moco domain, and a C-terminal dimerization domain. It is located in the mitochondrial intermembrane space of mammalian cells, where it aerobically converts sulfite to sulfate while concomitantly reducing two molecules of cytochrome c, representing the last step in the enzymatic catabolism of cysteine. Mechanistic studies indicate that during the sulfite-SO-cytochrome c catalytic cycle, movement between the molybdenum and heme domain is required to enable efficient single-electron transfer from molybdenum via the heme b5 cofactor to cytochrome c (21).
Since nitrite-dependent NO synthesis is enhanced under hypoxia, we hypothesized that under hypoxic or ischemic conditions a fully reduced system would serve to switch sulfite-mediated cytochrome c reduction in mitochondria to a nitrite reduction reaction. Here, we report that SO can function as source of bioactive NO by reducing nitrite at the molybdenum domain, coupled to a one-electron oxidation of Mo(IV)-to-Mo(V).
Results
Reduced human SO converts nitrite to NO
To determine whether prereduced SO reduces nitrite to NO at physiological concentrations of nitrite (8, 14), we performed measurements in a purge vessel under anaerobic conditions (helium gas purging through the reaction mixture). Rates were determined by integration of the peak NO generation rate over a 12-second interval (detailed methods are provided in the Supplementary Data; Supplementary Data are available online at www.liebertpub.com/ars). We validated these measurements using a purge and nonpurge system with both fast (nitrite in acidified iodide) and slow (DETA-NONOate) NO donor standards and using SO holo enzyme preparations (Supplementary Figs. S1 and S2). We use the purge vessel system for our kinetic measurements, as this has been the standard in the field evaluating molybdenum enzymes as nitrite reductases (25–27, 51), allowing for comparisons of measured reaction kinetics. We appreciate some limitations using this method related to protein denaturation over time, exhaustion of the reducing agent, and potential oxidative damage to the Moco center.
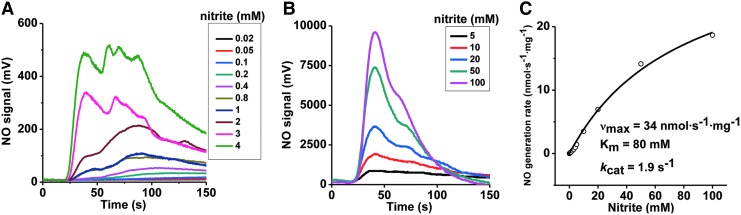
Our experiments used recombinant human SO ([Mo]=0.63 μM), and phenosafranine (one- or two-electron donor) or sulfite (two-electron donor) as reducing substrates. To study the kinetics of SO-dependent nitrite reduction, first the concentration dependence of nitrite was assessed in the presence of a fixed concentration of phenosafranine (40 μM) and human SO. A concentration-dependent increase in the rate of NO formation was observed for nitrite (0.02–100 mM) at pH 7.4 and fitted to the Michaelis–Menten equation (Fig. 1). When concentrations of nitrite were above 4 mM, the NO detected was more than 1000 mV at the high-sensitivity mode, and, thus, the low-sensitivity mode was set to measure NO production when nitrite was in the range of 5–100 mM. Measured vmax, Km, and kcat values are 34 nmol·s−1·mg−1, 80 mM, and 1.9 s−1, respectively. At pH 6.5, the rate of NO formation also increased with the concentration of nitrite, although the kinetic parameters were not determined (Supplementary Fig. S3).
FIG. 1.
Kinetics of NO generation from human holo-SO as a function of nitrite, in the presence of phenosafranine. Initial rates of NO generation were measured anaerobically at pH 7.4, 37°C, in 20 mM BisTris buffer. (A) NO generation traces with 0.63 μM SO and 40 μM reduced phenosafranine in the presence of 20 μM–4 mM nitrite at pH 7.4. High sensitivity mode was used in the range of 20 μM–4 mM. (B) Similar measurements were performed as (A) at low sensitivity mode in the range of 5–100 mM nitrite. (C) Corresponding NO generation rates were calculated from the representative traces shown in (A, B). NO, nitric oxide; SO, sulfite oxidase.
It is notable that SO only catalyzes a single turnover reaction in the presence of sulfite, with no steady-state NO formation observed. However, the relationship between the observed rates and the concentration of nitrite shows a noticeable deviation from a lineal relationship and appears to indicate a possible reaction complex formation. We interpret this fact as the result of a reaction as follows:
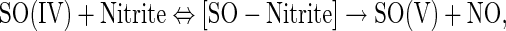 |
where SO(IV) and SO(V) represent SO proteins with their molybdenum atom in the oxidation states IV and V, respectively (25, 27). This kind of single turnover reaction can produce kobs versus [substrate] plots similar to Michaelis–Menten (17). We have renamed the calculated parameters Kd and ket, to indicate that they correspond to an apparent dissociation constant and an electron transfer rate for reactions in the presence of sulfite.
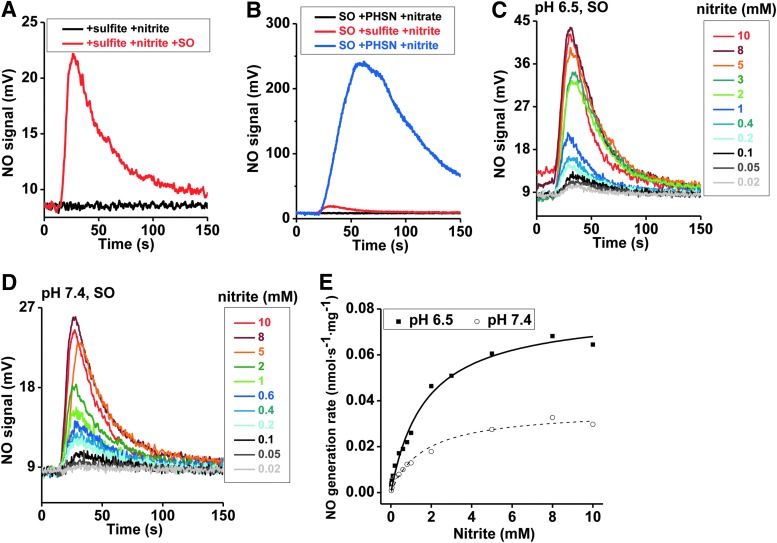
Since sulfite is the only known physiological reducing substrate for SO, NO generation was next studied with sulfite as the reductant. In the absence of SO or in the presence of only sulfite or nitrite, NO formation was not detected; while the combination of nitrite and SO in the presence of either sulfite or phenosafranine resulted in significant NO generation, suggesting redox coupling of substrate oxidation to nitrite reduction (Fig. 2A, B). It is important to note that the addition of nitrate does not lead to NO formation, suggesting that SO is not a nitrate reductase. NO generation with phenosafranine is ∼10-fold greater than that observed with sulfite (Fig. 2B), indicating that phenosafranine is a more efficient electron donor for nitrite reduction. This different reactivity with a strict two-electron donor, sulfite, versus a one- or two-electron donor such as phenosafranine prompted us to study the reaction mechanism in greater detail, as discussed next.
FIG. 2.
Representative traces and kinetics of NO generation from human holo-SO, in the presence of sulfite. Initial rates of NO generation were measured anaerobically at pH 6.5 and 7.4, 37°C, in 20 mM BisTris buffer. (A) Traces show NO generation from the reaction of nitrite with sulfite, in the absence (black) or in the presence of SO (red), with SO (1.5 μM), nitrite (1 mM) and sulfite (10 μM) at pH 6.5. (B) Traces show NO generation by 0.63 μM SO reacting with various reductant and oxidant combinations at pH 6.5: sulfite+nitrite (red), phenosafranine (PHSN)+nitrate (black), and PHSN+nitrite (blue). Concentrations of PHSN, sulfite, nitrate, and nitrite are 40 μM, 40 μM, 1 mM, and 1 mM, respectively. (C) NO generation traces with 0.63 μM SO and 5 μM sulfite in the presence of 20 μM–10 mM nitrite at pH 6.5. (D) NO generation traces at 7.4. (E) Corresponding NO generation rates were calculated from the representative traces shown in (C, D).
To study the kinetics of SO-dependent nitrite reduction, first the concentration dependence of nitrite was assessed in the presence of a fixed concentration of sulfite (5 μM) and human SO. A concentration-dependent increase in the rate of NO formation was observed for nitrite (0.02–10 mM), and a plot of kobs versus nitrite indicated a hyperbolic fit similar to Michaelis–Menten kinetics for both pH 6.5 and 7.4 (Fig. 2C–E), with vmax, Kd, and ket values of 0.0361 nmol·s−1·mg−1, 1.6 mM and 0.002 s−1 at pH 7.4 and 0.0796 nmol·s−1·mg−1, 1.7 mM and 0.0044 s−1 at pH 6.5, respectively. The increases of vmax and ket with the lower pH are not unexpected based on Equation 1 and have been reported for the nitrite reduction reactions catalyzed by hemoglobin, myoglobin, and neuroglobin (10, 40, 46). However, the NO generation rates only increased for about three-fold per unit change of pH, instead of the well-known 10-fold increase for a reaction of heme globins with nitrous acid. The reasons for different influences of pH, which was also observed in other Mo-dependent enzymes (25–27), are unknown but may involve a direct binding of nitrite or protonation of active site residues at the Mo center. As discussed and evaluated next, the low ket values using sulfite as the reductant suggest that this may not be a catalytic reaction.
When sulfite concentration was examined for effects on NO formation, no significant differences in the rate of nitrite reduction to NO were observed (2 μM–5 mM, pH 6.5) (Supplementary Fig. S4A). However, at pH 7.4, NO formation rates were decreased at sulfite concentrations greater than 1 mM, suggesting a substrate-mediated inhibitory effect on nitrite reduction (Supplementary Fig. S4B). These results suggest that both nitrite and sulfite react at the molybdenum active site. Finally, all results were reproduced with mouse SO (Supplementary Table S1 and Supplementary Fig. S4C–H).
Nitrite reduction to NO at the molybdenum domain of SO
To further assess whether nitrite is reduced at the molybdenum (Mo) center, we next evaluated reactions of nitrite with isolated recombinant Mo- and heme-domains of human SO. Recombinant Mo-domain was used for kinetic studies of NO generation as a function of nitrite or phenosafranine or sulfite concentration, and for quantification of nitrite reduction in the presence of phenosafranine or sulfite.
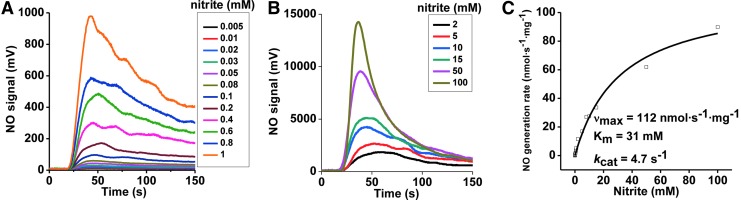
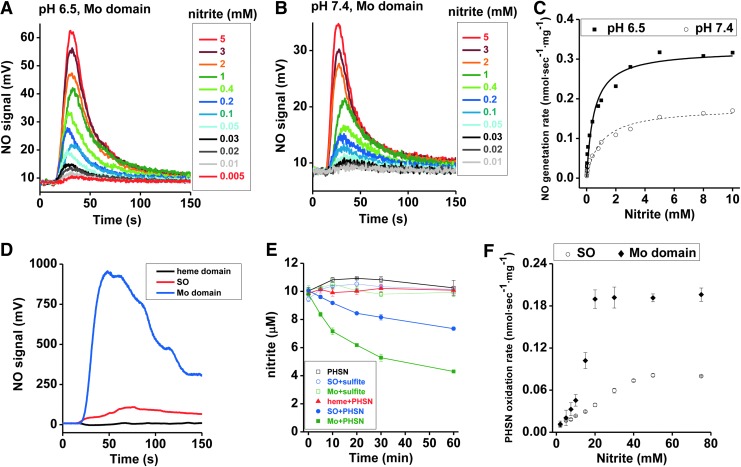
The concentration dependence of nitrite was first determined at a fixed concentration of 40 μM phenosafranine. On addition of 0.25 μM Mo-domain to nitrite and phenosafranine, NO formation was measureable with the lowest detectable NO concentration at 5 μM nitrite at pH 7.4 (Fig. 3A, B). Michaelis–Menten kinetics were obtained showing vmax, Km, and kcat values with 112 nmol·s−1·mg−1, 31 mM, and 4.7 s−1 at pH 7.4, respectively, with significantly higher vmax and kcat, and lower Km values than observed with the holo-enzyme (Supplementary Table S1). At pH 6.5, when nitrite was in the range of 1–400 μM, NO production by Mo-domain was faster than at pH 7.4 (Supplementary Fig. S5). When sulfite was used as the reducing substrate, the rates showed a similar dependence toward nitrite but were much lower than those observed in the presence of phenosafranine. The rates observed at pH 6.5 were higher than those at pH 7.4, as observed in the presence of phenosafranine (Fig. 4A–C), with vmax, Kd, and ket values as 0.179 nmol·s−1·mg−1, 0.973 mM and 0.0075 s−1 at pH 7.4, and 0.323 nmol·s−1·mg−1, 0.496 mM and 0.0136 s−1 at pH 6.5.
FIG. 3.
Kinetics of NO generation from Mo-domain as a function of nitrite, in the presence of phenosafranine. Initial rates of NO generation were measured anaerobically at pH 7.4, 37°C, in 20 mM BisTris buffer. (A) NO generation traces with 0.25 μM SO and 40 μM reduced phenosafranine in the presence of 5 μM–1 mM nitrite at pH 7.4. High sensitivity mode was used in the range of 5 μM–1 mM. (B) Similar measurements were performed as (A) at low sensitivity mode in the range of 2–100 mM. (C) Corresponding NO generation rates were calculated from the representative traces shown in (A, B).
FIG. 4.
NO generation and nitrite reduction by various types of engineered SO proteins. (A, B) Anaerobic NO generation by Mo-domain (0.25 μM) as a function of nitrite concentration (5 μM–5 mM) in the presence of 5 μM sulfite, at pH 6.5 (A) and 7.4 (B), 37°C, in 20 mM BisTris buffer. (C) Corresponding NO generation rates were calculated from the representative traces shown in (A, B). (D) Anaerobic NO generation from the reaction of nitrite (1 mM) with the Mo-domain, holo-SO, or heme domain ([Mo]=0.25 μM for Mo-domain; [Mo]=0.63 μM for holo-SO; [heme]=0.63 μM) at pH 7.4, 25°C, 20 mM BisTris buffer, with 40 μM PHSN was the reducing substrate. (E) Nitrite (10 μM) reduction is carried out in the anaerobic glove box at room temperature when nitrite reacts with SO+sulfite (open circle, blue), SO+PHSN (dark circle, blue), Mo-domain+sulfite (open square, green), Mo-domain+PHSN (dark square, green), heme domain+PHSN (dark triangle, red), and PHSN (open square, black). Concentrations of sulfite, PHSN, SO, Mo-domain, and heme domain are 50, 50, 10, 10, and 10 μM, respectively. (F) Steady-state kinetics of nitrite reduction by PHSN catalyzed by human holo-SO and the isolated Mo-domain, recorded spectrophotometrically at 520 nm with at least triplicate measurements. 12 μM human holo SO or human SO Mo-domain was mixed with 250 μM phenosafranine and 0–75 mM nitrite. The reaction was started with 250 μM sodium dithionite, and control experiments were performed without the addition of enzyme.
Our studies using phenosafranine or sulfite as reducing substrate show much faster nitrite reduction to NO by the Mo-domain than by holo-SO (Supplementary Table S1). Mechanistically, these data suggest that nitrite reacts with the Mo-domain, and that electron transfer between Mo- and heme domains and/or steric hindrance of the heme domain reduces the reaction rates with nitrite by approximately a factor of three. All results could be reproduced with the mouse Mo-domain using sulfite as the reductant (Supplementary Figs. S6 and S7).
To investigate the inhibitory effect of the heme domain, we evaluated rates of NO formation and nitrite reduction in the presence of the heme domain, the Mo-domain, and holo-SO at the same concentrations (Fig. 4D, E). When 40 μM phenosafranine was mixed with 1 mM nitrite in an anaerobic vessel, addition of the isolated heme domain did not generate NO; whereas addition of the Mo-domain and holo-SO resulted in NO formation (Fig. 4D), suggesting that the heme domain alone does not reduce nitrite to NO. When 10 μM nitrite was mixed with 50 μM sulfite under anoxic conditions, nearly zero nitrite was reduced in 60 min in the presence of holo SO or the Mo-domain (Fig. 4E). However, when 50 μM phenosafranine replaced sulfite, around 25% and 55% nitrite were reduced anaerobically in the presence of holo-SO and the Mo-domain, respectively, while the heme domain did not reduce nitrite (Fig. 4E). Furthermore, nitrite-mediated phenosafranine oxidation by both the Mo-domain and holo-SO confirmed that the catalytic activity of the Mo-domain is ∼2.5-fold greater than observed with the holoenzyme (Fig. 4F). All these results provide further evidence that nitrite reduction occurs at the Mo-domain, with enhanced reaction rates in the presence of phenosafranine compared with sulfite.
Effects of the heme domain on rates of nitrite reduction
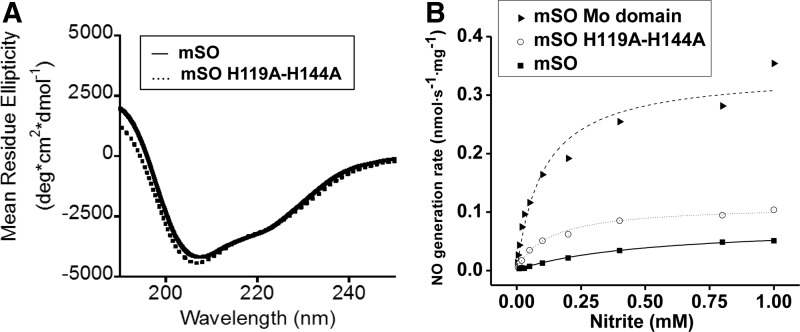
To determine whether either the heme-iron center or the heme domain itself negatively modulates NO production within the holoenzyme, we quantified the NO generation rates in the presence of a heme-deficient mouse SO, H119A/H144A variant (Fig. 5 and Supplementary Fig. S8A–C). Loss of His119 and His144, which coordinate heme iron, results in an apo-heme domain, with an overall preservation of the enzyme structure as confirmed by circular dichroism (CD) spectroscopy (Fig. 5A). Functionally, the H119A/H144A SO and holo-SO exhibited ∼30% and 20% of the NO formation activity of the isolated Mo-domain, respectively, suggesting that there is both a steric and an electronic inhibitory effect from the heme domain on Mo-domain-dependent nitrite reduction (Fig. 5B and Supplementary Fig. S8A–C).
FIG. 5.
Circular dichroism (CD) spectroscopy of wild-type mouse SO and H119A/H144A and NO generation by mouse SO proteins. (A) CD spectra of the secondary structure of wild-type mouse SO (—, solid line), H119A/H144A (…, dot line). (B) NO generation rates as a function of nitrite concentration catalyzed by wild-type mouse SO, molybdenum domain of mouse SO, and the H119A/H144A mouse SO mutant. SO (—, solid line, square), H119A/H144A (…, dot line, open circle), and Mo domain (—, dash line, triangle).
Catalytic Mo(IV) to Mo(V) nitrite reduction cycle
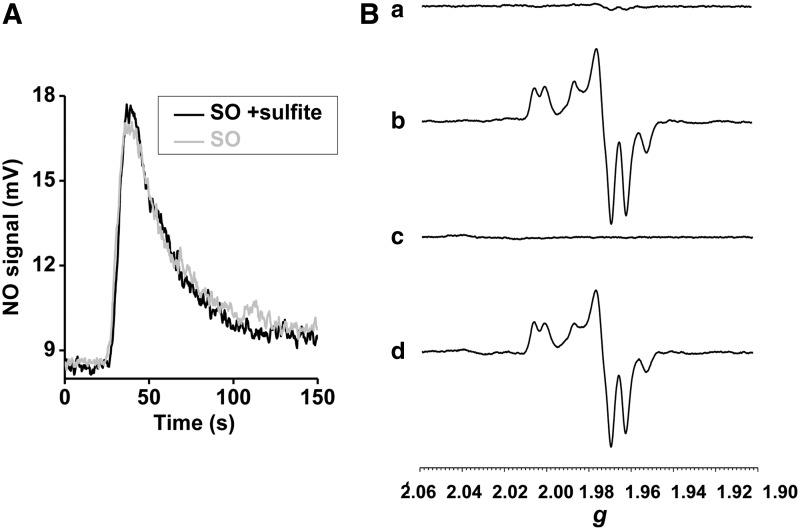
Based on the observation that phenosafranine, a one- or two-electron donor, efficiently catalyzes nitrite reduction by SO, while sulfite (a strict two-electron donor) does not, we studied reactions of prereduced SO with nitrite under anaerobic conditions, in the presence or absence of sulfite. Importantly, addition of sulfite did not promote additional NO generation beyond that of the fully prereduced SO (Fig. 6A). This result suggests that sulfite cannot enter into a catalytic cycle with nitrite reduction, that is, sulfite only modulates one turnover of nitrite reduction. On the other hand, addition of phenosafranine to reduced SO increased NO production, suggesting that a one-electron donor can catalyze nitrite reduction by SO, forming a redox cycle of phenosafranine oxidation and nitrite reduction.
FIG. 6.
NO generation by prereduced human holo-SO; EPR spectroscopy of the human Mo-domain. (A) Anaerobic NO generation rates with 0.63 μM prereduced human SO and 1 mM nitrite, in the presence or absence of 5 μM sulfite at pH 6.5, 37°C, in 20 mM BisTris buffer. (B) Anaerobic EPR spectra of the Mo center in the isolated Mo-domain of human SO. (a) 20 μM Mo-domain+100 μM sulfite, (b) 1 mM nitrite was added to (a), (c) 50 μM reduced PHSN was added to (b), (d) thawing and incubation of (c) for 1.5 h. EPR, electron paramagnetic resonance.
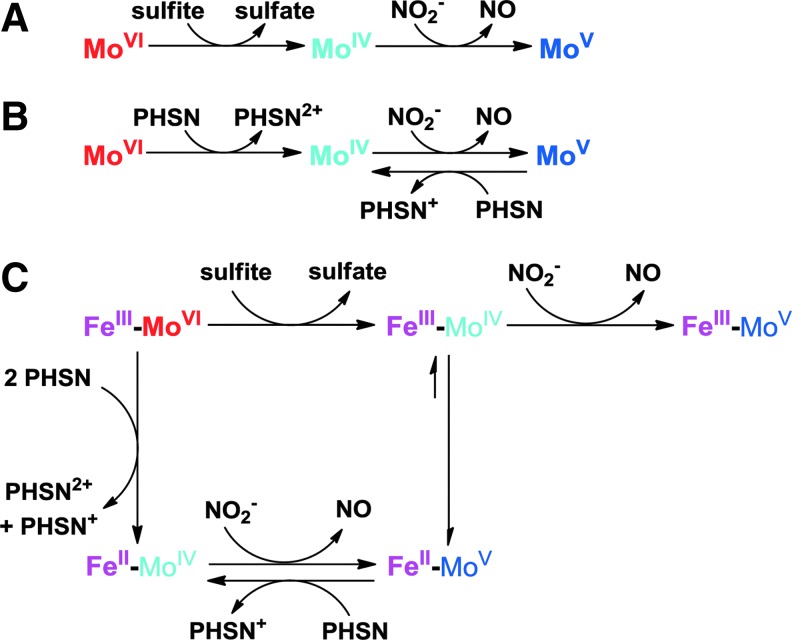
We applied electron paramagnetic resonance (EPR) spectroscopy to monitor changes at the Mo center during its reaction with sulfite, nitrite, and phenosafranine. When 20 μM Mo-domain was mixed with 100 μM sulfite, the Mo center was EPR silent, reflecting the anticipated reduced Mo(IV) state (Fig. 6B-(a)). When excess nitrite (1 mM) was added and incubated for 1.5 h, a stable Mo(V) spectrum appeared (23) and no further change was observed (Fig. 6B-(b)), indicating a one-electron oxidation but inability to convert to the Mo(VI) oxidation state. On addition of 50 μM reduced phenosafranine to the solution and quick freezing afterward (15 s), the Mo(V) spectrum disappeared, suggesting a conversion of Mo(V) back to the EPR-silent Mo(IV) (Fig. 6B-(c)). On thawing and incubation with original added nitrite (1 mM) again for another 1.5 h, more than 80% of the Mo(V) signal was recovered (Fig. 6B-(d)). These data suggest that Mo(IV), formed initially from the two-electron reduction of Mo(VI) by sulfite, is then oxidized to Mo(V) by nitrite (model shown in Fig. 7). Importantly, nitrite cannot further oxidize Mo(V) to EPR-silent Mo(VI). Since sulfite cannot reduce Mo(V) to Mo(IV), the reaction stops at Mo(V). Addition of phenosafranine is able to reduce Mo(V) to Mo(IV), enabling nitrite to be again reduced (Fig. 7). These data, in aggregate, explain the lack of redox cycling of nitrite and sulfite and evident catalytic cycling of nitrite and the one-electron donor phenosafranine (Fig. 7).
FIG. 7.
Proposed electron transfer in the Mo-domain and holo-SO. Proposed electron transfer between the Mo center and (A) sulfite or nitrite, (B) PHSN or nitrite; (C) proposed electron transfer between the two metal centers, Mo and heme-Fe, and a substrate (sulfite, nitrite, or PHSN).
SO catalyzes nitrite-NO-cGMP signaling in human fibroblasts
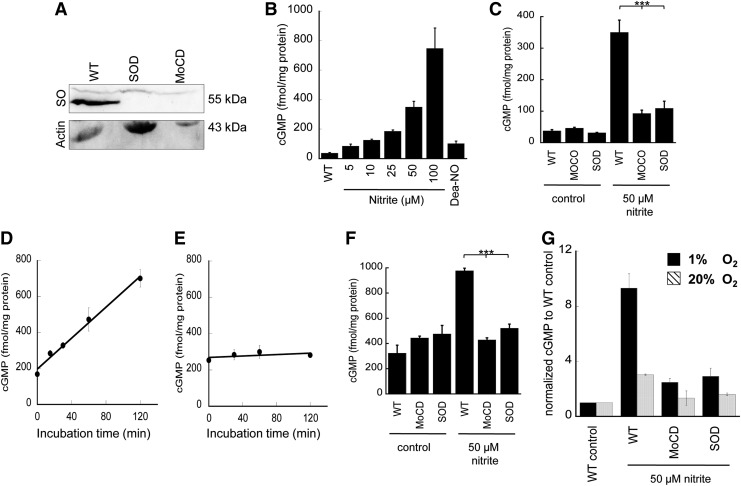
Fibroblasts isolated from patients with Moco deficiency (MOCD), which produces inactive SO, xanthine oxidase, and aldehyde oxidase, as well as fibroblasts from an SO-deficient (SOD) patient were used to study hypoxic nitrite exposure on cGMP generation. Control experiments were performed using fibroblasts from healthy individuals cultured and treated under identical hypoxic conditions (1% O2). SO expression in MOCD or SOD fibroblasts was not detectable when 120 μg protein was loaded for Western analysis assay (Fig. 8A), indicating the deficiency of SO in patients' cells.
FIG. 8.
Expression of SO in human fibroblast and cGMP formation from human fibroblast. (A) SO expression in fibroblasts from healthy individuals (WT), SO-deficient (SOD) or Moco-deficient (MOCD) patients. 120 μg protein from the cell lysate was loaded for electrophoresis. (B) Human fibroblast cells derived from WT were treated with 1 μM sildenafil, and then with various concentrations of nitrite or 5 μM DEA-NO followed by 2 h hypoxic incubation (1% O2). Cyclic GMP (cGMP) levels were determined in cell lysates. Data represent mean±SD. (C) Human fibroblast cells derived from SOD or MOCD patients or WT were treated with 1 μM sildenafil, and then with 50 μM nitrite followed by 2 h of hypoxic incubation (1% O2). Control samples are without nitrite treatment. (***p<0.001, n= 3). (D) Human fibroblasts derived from WT were treated with 1 μM sildenafil, and subsequently with 100 μM nitrite followed by hypoxic incubation for 0, 15, 30, 60, or 120 min. cGMP levels were determined accordingly (y=4.296x+197.52, R2=0.995). (E) The same kinetics of cGMP formation as shown in (D) was carried out in SO-deficient fibroblasts (y=0.199x+268.86, R2=0.533). (F) Human fibroblast cells derived from SOD or MOCD patients or WT were treated with 1 μM sildenafil, and then with 50 μM nitrite under 2 h normoxia (20% O2). Control samples are without nitrite treatment. (***p<0.001, n=3). (G) Comparison of cGMP production in (C) and (F) after normalized to the WT control. DEA-NO, DEA-NONOate; Moco, molybdenum cofactor; MOCD, molybdenum cofactor deficiency; WT, wild type.
Control experiments were performed using fibroblasts from healthy individuals cultured and treated under identical hypoxic conditions (1% O2). In the presence of an NO donor, DEA-NONOate (DEA-NO), cGMP levels significantly increased (Fig. 8B). Addition of nitrite (5–100 μM) to the medium produced a dose-dependent increase in cellular cGMP (Fig. 8B). For fibroblasts from either Moco- or SO-deficient patients treated with 50 μM nitrite, the cGMP generation was reduced by approximately 80% (Fig. 8C), indicating a classic nitrite-NO-soluble guanylyl cyclase-cGMP response. A time-dependent exposure of wild-type fibroblasts to nitrite showed a linear increase of cGMP levels over time (120 min) with a rate of 4.3 fmol cGMP per mg and minute (Fig. 8D). In contrast, SO-deficient cells showed no significant cGMP production (Fig. 8E) in the same time frame. Similar exploration of cGMP production was performed under normoxia (20% O2). The basal level of cGMP at 20% O2 is higher than at 1% O2 (Fig. 8F), which indicates higher oxygen availability for NO synthase-dependent NO formation in fibroblasts under normoxia. Treatment with nitrite increases cGMP production in fibroblasts even at 20% oxygen, but the fold increase was less under normoxia compared with hypoxia (Fig. 8G), which is also supported by less NO generation under normoxia detected in the NO analyzer (Supplementary Fig. S9). Most importantly, the effect of nitrite on cGMP formation was abrogated in the Moco- or SO-deficient patient's fibroblasts (Fig. 8F). These results demonstrate that SO represent the primary Moco-dependent enzyme required for enzymatic conversion of nitrite to NO in human fibroblasts, given that the observed nitrite-dependent cGMP formation is impaired in SO-deficient cells.
Discussion
Mammals utilize two pathways to produce NO and regulate blood pressure and blood flow: the L-arginine-to-NO pathway and the nitrate-to-nitrite-to-NO pathway (29). These NO biosynthesis pathways respond differently to oxygen tension. The arginine-to-NO pathway requires oxygen as a substrate for NO formation; conversely, the nitrate-to-nitrite-to-NO pathway is potentiated under hypoxic conditions. The upstream pathways for conversion of nitrate to nitrite have been clearly defined, as earlier studies first described the involvement of this pathway in the generation of NO in the stomach, a process shown to be important in regulating stomach mucosal integrity, mucosal host defense, and later mucosal blood flow (5, 6, 28). Since the nitrate-to-nitrite-to-NO pathway exhibits increased potency under lower oxygen tensions, a role for nitrite as an effector of hypoxic signaling and vasodilation has been considered (10, 16, 29). While aerobic NO production has been extensively investigated, enzymes that regulate nitrite-to-NO reduction are less well understood.
Several studies have investigated the possible enzymatic origins for mammalian nitrate and nitrite reduction, and a number of enzyme systems have been reported to control nitrite reduction to NO under certain conditions. As mentioned, xanthine oxidase has been proposed as a mammalian nitrate and nitrite reductase (18). A number of studies have demonstrated that the therapeutic effects of nitrite on pulmonary hypertension require a functional xanthine oxidase system (2, 55); however, the inhibition of xanthine oxidase does not inhibit nitrite-dependent vasodilation in human circulation (12). Several other mammalian nitrite reductase candidates have been proposed, such as the other Mo-containing enzyme aldehyde oxidase and heme enzymes (cytochrome c, eNOS, deoxyhemoglobin, deoxymyoglobin, neuroglobin, and cytoglobin), as well as one nitrite anhydrase (carbonic anhydrase) (1, 4, 19, 38, 40). However, it is likely that additional catalytic human nitrate and nitrite reductases have not yet been identified, such as SO, which has comparable or higher activity than xanthine oxidase and aldehyde oxidase when phenosafranine, a one- or two-electron donor, was used as the reducing substrate. When sulfite, a strict two-electron donor and a known physiological substrate of SO, was used as the reductant, less NO was produced.
These studies suggest that reduced SO can reduce nitrite to form NO, in a reaction analogous to that of xanthine oxidase and aldehyde oxidase. It is now clear that many heme-based and molybdopterin-based proteins have the capacity to reduce nitrite to NO. Enzymes that metabolize nitrite to NO are likely dependent on specific organ location and local oxygen tensions. For example, deoxyhemoglobin reduces nitrite to NO in the blood, and this pathway has been shown to regulate hypoxic vasodilation (10, 11) and, more recently, inhibition of platelet activation (42). Deoxymyoglobin plays a major role in the heart (41, 48) and smooth muscle, where it regulates mitochondrial respiration, myocardial energetics, and hypoxic vasodilation. Consistent with a role for heme globins in vasodilatory and platelet inhibitory effects of nitrite, oxypurinol does not inhibit nitrite-dependent vasodilation.
On the other hand, treatment of rodents with tungsten, which replaces molybdenum in xanthine oxidase, aldehyde oxidase, and SO, inhibits the protective effects of nitrite in vascular injury models, such as carotid wire injury and pulmonary arterial hypertension (2, 54). Allopurinol can also inhibit the effects of nitrite in many preclinical disease models (15). Our present studies using fibroblasts for SO-deficient patients suggest that SO plays a dominant role in nitrite reduction and sGC activation in these cells. These data, in aggregate, suggest that a number of enzymes can reduce nitrite to NO, and the dominant effect of one system likely depends on enzyme concentration and oxygen concentration in specific tissues. Future work with conditional knockout of xanthine oxidase, aldehyde oxidase, and SO will be required to fully explore the relative importance of each in specific tissues and diseases.
In this study, we examined the enzymatic reactions of human and mouse SO in vitro with phenosafranine, sulfite, and nitrite using NO detection, nitrite reduction, and EPR methodologies, revealing that SO can catalyze the reduction of nitrite to NO by the one-electron oxidation of Mo(IV) to Mo(V). We unequivocally demonstrated that only fully reduced Mo(IV) enzyme is capable of nitrite reduction, while Mo(V) does not produce NO. This result indicates that both the electron donor and the redox potential of the enzymatic metal center play an important role in catalytic reactions, which determines the electron flow in SO. SO has two internal cofactors, heme and molybdenum, which have reduction potentials of 68 mV (heme), −239 mV (Mo(V)/Mo(IV)), and 38 mV (Mo(VI)/Mo(V)), respectively (9). Electrons flow from the Mo center to the heme center, and Mo(V) is the favorable oxidation state. In order to efficiently catalyze nitrite reduction in vivo, either an appropriate one-electron donor can reduce Mo(V) to Mo(IV) or an oxidant can oxidize Mo(V) to Mo(VI), enabling sulfite to react with SO again. After the nitrite-dependent formation of Mo(V), the remaining electrons may be transferred to the heme domain, which, in turn, requires one cycle of cytochrome c reduction to complete the catalytic cycling.
Given that reduced SO was not found to react with nitrate, we conclude that SO cannot function as nitrate reductase. A recent study by Qiu et al. has engineered an SO enzyme variant with partial nitrate reductase activity using structure-guided mutagenesis (35). Plant nitrate reductases have three internal cofactors: FAD, heme, and molybdenum cofactor (Moco). Their reduction potentials are −272 to −187 mV, −123 to −174 mV, and −25 to +15 mV, respectively (9). This redox pattern is consistent with a downhill flow of electrons within the enzyme, that is, electrons are readily transferred from heme to Mo for nitrate reduction. In addition, electrons from NAD(P)H, the physiological reductant, are passed to heme and Mo sites one by one via the FAD domain. However, the well-accepted flow of electrons in SO is from the Mo domain to the heme domain, and finally to cytochrome c, the physiological oxidant.
Our studies also suggest that the heme domain reduces the rate of nitrite reduction to NO. The fact that the apo-heme domain also inhibits nitrite reduction, to some extent, suggests an important conformational component in the access of nitrite to sulfite-reduced SO. There is a flexible hinge (14 residues in human SO) that tethers N-terminal heme domain to Mo-domain (21). Previous studies have shown that this hinge controls domain motion during the catalytic cycle of SO, which adopts various conformations during intra-molecular electron transfer (IET) and catalysis (49). Thus, domain motion appears to change domain distance and modulate activity, as supported by molecular dynamic studies that propose a much shorter Mo-heme distance (19 Å) during IET as compared with the distance in the crystal structure of SO (32 Å) (37). Electronically, there are six possible redox states of SO, depending on oxidation states of molybdenum and heme iron. Under hypoxic, ischemic, or reductive conditions, cytochrome c is fully reduced, which, in turn, will slow down re-oxidation of the heme iron and, ultimately, inhibit IET between Mo and heme. This can provide a possible mechanism for hypoxic and/or ischemic nitrite-dependent NO formation catalyzed by SO.
In addition to hypoxia, sulfite accumulation would increase the steady-state levels of Mo(IV)-reduced SO. However, such a condition is only known for Moco- or SO deficiency, genetic disorders leading to rapidly neurodegeneration, mental retardation, and early childhood death. Recently, a first successful experimental treatment for MOCD has been reported (50). Those patients lack Moco and, consequently, sulfite accumulates in large quantities. Due to the reconstitution of Moco biosynthesis in treated patients, SO activity returns and excess sulfite is removed with 72–96 h post-treatment (50). In this study, Moco- or SO deficiency in isolated human fibroblasts caused significant decreases of nitrite-dependent cGMP generation, supporting a role for SO in nitrite-dependent intracellular NO signaling.
This work has identified human SO as a new candidate nitrite reductase that leads to NO production both in vitro and in vivo, which requires a one-electron donor to drive the catalytic cycle between Mo(IV) and Mo(V) in the purified enzyme system. SO contributes to NO-soluble guanylyl cyclase-cGMP signaling in human fibroblasts. Further studies are needed to elucidate the physiological mechanism and significance of how SO function in nitrite-NO signaling in human cells and tissues.
Materials and Methods
Reagents and standard samples preparation
All chemicals were purchased from Sigma-Aldrich. Holo SO, isolated Mo, and heme domains were reduced by sulfite or dithionite; purified by passing through a Sephadex G-25 gel filtration column; and eluted with the indicated buffer anaerobically. Concentrations of the heme cofactor in holo SO and heme domain were determined by measuring absorbance of the heme Soret band using ɛ414=113 mM−1·cm−1 or ɛ423=164 mM−1·cm−1. Concentrations of the Mo cofactor in holo SO and isolated Mo-domain were determined by HPLC Form A analysis (22).
Cloning, expression, and purification of the recombinant proteins
Mature human and mouse SO were polymerase chain reaction cloned into the SacI and SalI sites of pQE-80L (Qiagen). The resulting constructs were used to create the Mo and heme domain expression constructs, as well as the heme-deficient variant (H119A/H144A) using the same restriction sites. All constructs were expressed in Escherichia coli strain TP1000 (33, 46) for 48 h at 24°C. Protein expression from pQE80 was induced after 4 h of incubation at 24°C with 100 μM IPTG. 500 μM sodium molybdate was added to the culture to achieve sufficient levels of Moco production. His-tagged SO was affinity purified using Ni-NTA resin (Qiagen) according to the manufacturer's instructions. Protein purification included further anion exchange with a SourceQ15 column (GE Healthcare) using 50 mM Tris-acetate, pH 8.0, as buffer A. Proteins were eluted with a gradient from 100 to 500 mM NaCl in buffer A.
Measurement of NO formation by ozone-based chemiluminescence
Detailed methods are provided in the Supplementary Data. In summary, NO production was measured by ozone-based chemiluminescence using a Sievers Nitric Oxide Analyzer (NOA 280i) equipped with a liquid sampling purge system (GE Analytical Instruments). Sievers NO Analysis™ Software, Liquid (Version 3.2) was used for data acquisition, at a rate of one data point per 0.25 s.
The standard glass purge vessel (15-ml) was used to mix and deoxygenate reagents. For enzyme reactions, the reaction buffer (20 mM BisTris buffer or 10 mM phosphate buffer pH 6.5 or 7.4) was maintained in the presence of nitrite for 2 min in the sealed purge vessel to equilibrate the temperature to 37°C and to void oxygen from the reagents before initiating the enzyme reaction. Next, reducing substrate (sulfite or phenosafranine) was injected into the purge vessel through the vessel's septa using a Hamilton syringe. Once a stable baseline was established, SO was injected into the mixture and the reaction was monitored for several minutes, until the reaction was completed.
NO production rates were estimated using a calibration curve prepared by the nitrite/I3− method (see Supplementary Data for details). All the NO generation rates were plotted against the corresponding concentrations of nitrite, and fit to a hyperbolic equation, y=(P1×x)/(P2+x). The values of P1 and P2 correspond to νmax and Km values in the Michaelis–Menten equation, that is, ν=Vmax×[S]/(Km+[S]). In the case of single turnover reactions (nonsteady state), a similar fit was used, where the P1 and P2 terms represent the apparent νmax and Km values, which we refer to as ket (rate of electron transfer) and Kd (apparent dissociation constant) to indicate the single turnover nature of the studied reaction.
Nitrite reduction assay
Time-course nitrite reduction was evaluated by measuring the concentration of nitrite in the reaction mixture with the NOA 280i. 10 μM SO or isolated Mo domain was mixed with 10 μM nitrite, in the presence of sulfite (50 μM) or reduced phenosafranine (50 μM) inside the anaerobic glove box. Afterward, nitrite concentration was measured at 5, 10, 20, 30, and 60 min. Time zero was defined as the same reaction in the absence of protein. Each data point represents the average of three separate measurements.
Steady-state kinetics of nitrite reduction by phenosafranine
Steady-state kinetics were performed anaerobically in a glove box using a plate reader (Biotek). All data recorded are mean values of at least triplicate measurements. Initial velocities were determined by following the oxidation of the artificial electron donor phenosafranine at 520 nm using an extinction coefficient of 29,000 M−1·cm−1. SO was assayed at room temperature in Bis-Tris/acetate buffer, pH 6.5. In the phenosafranine-dependent reduction of nitrite, 12 μM human holo SO or human SO Mo-domain was mixed with 250 μM phenosafranine and 0–75 mM nitrite. The reaction was started with 250 μM sodium dithionite, and control experiments were performed without the addition of enzyme.
CD spectroscopy
Structural similarity between recombinant wild-type mouse SO and the heme-deficient H119A/H144A variant was determined by CD spectroscopy with a JASCO J-715 CD spectropolarimeter. All measurements were performed at 20°C, and secondary structure was measured over a wavelength range of 180–250 nm.
EPR spectroscopy
EPR spectra were recorded at 150 K with the EWWIN 6.0 acquisition and lineshape analysis software (Scientific Software Services) on a Bruker ER 300 spectrometer. Double integration was performed with EWWIN 6.0 using the “very rapid” xanthine oxidase Mo(V) signal as an integration standard (32). All reactions were performed in 50 mM Tris-acetate buffer, pH 8.0 anaerobically. In these experiments, sulfite (100 μM) was added to 20 μM anaerobic Mo domain; the reaction solution was transferred to an argon-flushed EPR tube and frozen; and an EPR spectrum was recorded. The sample was then thawed, 1 mM nitrite was added, and the mixture was incubated for 1.5 h at room temperature (RT) before being re-frozen and a second EPR spectrum was recorded. The sample was then re-thawed, 50 μM dithionite-reduced phenosafranine was added, the sample was re-frozen within 15 s of mixing, and a third EPR spectrum was recorded. Finally, the same solution was once again thawed, and the reaction mixture was incubated for an additional 1.5 h at RT before being frozen for the recording of a final EPR spectrum.
Western blot analysis
Fibroblast cells were washed with ice-cold PBS and homogenized in PBS with sonication. Then, 120 μg total protein of the cell lysate was loaded for electrophoresis. The cell lysates were resolved using 10% Bis-Tris SDS-PAGE gels and transferred to high-quality polyvinylidene difluoride (PVDF) membrane. The membrane was then probed with primary antibodies that were specific to SO (Eurogentec) and beta-actin (Santa Cruz). After incubation with the secondary antibodies coupled to horseradish peroxidase from Santa Cruz, the chemiluminescent signals were detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Determination of cGMP production in human fibroblasts
Human fibroblasts derived from SO- and Moco-deficient patients or healthy individuals were grown on 15 cm plates at 37°C with 5% CO2 in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 2 mM L-glutamine (Gibco). Cells were grown to about 90% confluence before treatment with 1 μM sildenafil and 5–100 μM nitrite or 5 μM DEA-NO for 2 h at hypoxic conditions (1% O2) or aerobically (20% O2). Control samples were incubated either without 5 μM DEA-NO or with nitrite addition. Time-dependent cGMP production was measured at 0, 15, 30, 60, and 120 min with 100 μM Nitrite. Cells were harvested and lysed with 100 μl HCl (0.1 M), and total protein concentration was determined by Bradford assay. Triplicates were used for each condition, and cGMP concentration was detected with a cGMP enzyme-linked immunosorbent assay kit (Cayman Chemicals). Results are expressed as the means±standard deviation. Statistical differences between two sets of data were calculated using a Student's t-test. Statistical significance was assumed when p was less than 0.001.
Supplementary Material
Abbreviations Used
- CD
circular dichroism
- DEA-NO
DEA-NONOate
- EPR
electron paramagnetic resonance
- IET
intra-molecular electron transfer
- Moco
molybdenum cofactor
- MOCD
molybdenum cofactor deficiency
- NO
nitric oxide
- SO
sulfite oxidase
- SOD
sulfite oxidase deficiency
- WT
wild type
Acknowledgments
The authors thank Dr. Jack Lancaster for helpful discussions on reaction kinetics in NOA purge systems. Dr. Gladwin receives research support from NIH grants R01HL098032, RO1HL096973, and PO1HL103455; the Institute for Transfusion Medicine; and the Hemophilia Center of Western Pennsylvania. Dr. Tejero acknowledges the support from the Competitive Medical Research Fund of the University of Pittsburgh Medical Center (UPMC) Health System. Dr. Guenter Schwarz thanks the German Research Foundation (DFG) and the Fonds der Chemischen Industrie for research support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aamand R, Dalsgaard T, Jensen FB, Simonsen U, Roepstorff A, and Fago A. Generation of nitric oxide from nitrite by carbonic anhydrase: a possible link between metabolic activity and vasodilation. Am J Physiol Heart Circ Physiol 297: H2068–H2074, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Alef MJ, Vallabhaneni R, Carchman E, Morris SM, Shiva S, Wang Y, Kelley EE, Tarpey MM, Gladwin MT, Tzeng E, and Zuckerbraun BS. Nitrite-generated NO circumvents dysregulated arginine/NOS signaling to protect against intimal hyperplasia in Sprague-Dawley rats. J Clin Invest 121: 1646–1656, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.This reference has been deleted
- 4.Basu S, Grubina R, Huang J, Conradie J, Huang Z, Jeffers A, Jiang A, He X, Azarov I, Seibert R, Mehta A, Patel R, King SB, Hogg N, Ghosh A, Gladwin MT, and Kim-Shapiro DB. Catalytic generation of N2O3 by the concerted nitrite reductase and anhydrase activity of hemoglobin. Nat Chem Biol 3: 785–794, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Benjamin N, O'Driscoll F, Dougall H, Duncan C, Smith L, Golden M, and McKenzie H. Stomach NO synthesis. Nature 368: 502, 1994 [DOI] [PubMed] [Google Scholar]
- 6.Bjorne HH, Petersson J, Phillipson M, Weitzberg E, Holm L, and Lundberg JO. Nitrite in saliva increases gastric mucosal blood flow and mucus thickness. J Clin Invest 113: 106–114, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.This reference has been deleted
- 8.Bryan N, Rassaf T, Maloney RE, Rodriguez CM, Saijo F, Rodriguez JR, and Feelisch M. Cellular targets and mechanisms of nitros(yl)ation: an insight into their nature and kinetics in vivo. Proc Natl Acad Sci U S A 101: 4308–4313, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell WH. Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Phys 50: 277–303, 1999 [DOI] [PubMed] [Google Scholar]
- 10.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu X, Huang KT, Shields H, Kim-Shapiro DB, Schechter AN, Cannon RO, and Gladwin MT. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat Med 9: 1498–1505, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Crawford JH, Isbell TS, Zhi Huang, Shiva S, Chacko BK, Schechter AN, Darley-Usmar VM, Kerby JD, Lang JD, Kraus D, Ho C, Gladwin MT, and Patel RP. Hypoxia, red blood cells, and nitrite regulate NO-dependent hypoxic vasodilation. Blood 107: 566–574, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dejam A, Hunter CJ, Tremonti C, Pluta RM, Hon YY, Grimes G, Partovi K, Pelletier MM, Oldfield EH, Cannon RO, Schechter AN, and Gladwin MT. Nitrite infusion in humans and nonhuman primates: endocrine effects, pharmacokinetics, and tolerance formation. Circulation 116: 1821–1831, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Dejam A, Hunter CJ, and Gladwin MT. Effects of dietary nitrate on blood pressure. N Engl J Med 356: 1590, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Feelisch M, Fernandez BO, Bryan NS, Garcia-Saura MF, Bauer S, Whitlock DR, Ford PC, Janero DR, Rodriguez J, and Ashrafian H. Tissue Processing of Nitrite in Hypoxia: an intricate interplay of nitric oxide-generating and -scavenging systems. J Biol Chem 283: 33927–33934, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh SM, Kapil V, Fuentes-Calvo I, Bubb KJ, Pearl V, Milsom AB, Khambata R, Maleki-Toyserkani S, Yousuf M, Benjamin N, Webb AJ, Caulfield MJ, Hobbs AJ, and Ahluwalia A. Enhanced vasodilator activity of nitrite in hypertension: critical role for erythrocytic xanthine oxidoreductase and translational potential. Hypertension 61: 1091–1102, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Gladwin MT, Shelhamer JH, Schechter AN, Pease-Fye ME, Waclawiw MA, Panza JA, Ognibene FP, and Cannon RO. Role of circulating nitrite and S-nitrosohemoglobin in the regulation of regional blood flow in humans. Proc Natl Acad Sci U S A 97: 11482–11487, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurley JK, Medina M, Gomez-Moreno C, and Tollin G. Further characterization by site-directed mutagenesis of the protein-protein interface in the ferredoxin/ferredoxin:NADP+ reductase system from Anabaena: requirement of a negative charge at position 94 in ferredoxin for rapid electron transfer. Arch Biochem Biophys 312: 480–486, 1994 [DOI] [PubMed] [Google Scholar]
- 18.Jansson EA, Huang L, Malkey R, Govoni M, Nihlen C, Olsson A, Stensdotter M, Petersson J, Holm L, Weitzberg E, and Lundberg JO. A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat Chem Biol 4: 411–417, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman T, Tejero J, Chen BB, Blood AB, Frizzell S, Shapiro C, Tiso M, Hood BL, Wang X, Zhao X, Conrads TP, Mallampalli RK, and Gladwin MT. 14-3-3 binding and phosphorylation of neuroglobin during hypoxia modulate six-to-five heme pocket coordination and rate of nitrite reduction to nitric oxide. J Biol Chem 286: 42679–42689, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.This reference has been deleted
- 21.Johnson-Winters K, Nordstrom AR, Emesh S, Astashkin AV, Rajapakshe A, Berry RE, Tollin G, and Enemark JH. Effects of interdomain tether length and flexibility on the kinetics of intramolecular electron transfer in human sulfite oxidase. Biochemistry 49: 1290–1296, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuper J, Llamas A, Hecht HJ, Mendel RR, and Schwarz G. Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430: 803–806, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Lamy MT, Gutteridge S, and Bray RC. Electron-paramagnetic-resonance parameters of molybdenum(V) in sulfite oxidase from chicken liver. Biochem J 185: 397–403, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen FJ, Schiffer TA, Borniquel S, Sahlin K, Ekblom B, Lundberg JO, and Weitzberg E. Dietary inorganic nitrate improves mitochondrial efficiency in humans. Cell Metab 13: 149–159, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Li H, Samouilov A, Liu X, and Zweier JL. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrite reduction. J Biol Chem 276: 24482–24489, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Li H, Cui H, Kundu TK, Alzawahra W, and Zweier JL. Nitric oxide production from nitrite occurs primarily in tissues not in the blood: critical role of xanthine oxidase and aldehyde oxidase. J Biol Chem 283: 17855–17863, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Kundu TK, and Zweier JL. Characterization of the magnitude and mechanism of aldehyde oxidase-mediated nitric oxide production from nitrite. J Biol Chem 284: 33850–33858, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundberg JO, Weitzberg E, Lundberg JM, and Alving K. Intragastric nitric oxide production in humans: measurements in expelled air. Gut 35: 1543–1546, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg JO, Weitzberg E, and Gladwin MT. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov 7: 156–167, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Lundberg JO. Nitrate transport in salivary glands with implications for NO homeostasis. Proc Natl Acad Sci U S A 109: 13144–13145, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maher AR, Milsom AB, Gunaruwan P, Abozguia K, Ahmed I, Weaver RA, Thomas P, Ashrafian H, Born GV, James PE, and Frenneaux MP. Hypoxic modulation of exogenous nitrite-induced vasodilation in humans. Circulation 117: 670–677, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Mcwhirter RB. and Hille R. The Reductive half-reaction of xanthine-oxidase -identification of spectral intermediates in the hydroxylation of 2-hydroxy-6-methylpurine. J Biol Chem 266: 23724–23731, 1991 [PubMed] [Google Scholar]
- 33.Palmer T, Santini CL, Iobbi-Nivol C, Eaves DJ, Boxer DH, and Giordano G. Involvement of the narJ and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol Microbiol 20: 875–884, 1996 [DOI] [PubMed] [Google Scholar]
- 34.Qin L, Liu XB, Sun QF, Fan ZP, Xia DS, Ding G, Ong HL, Adams D, Gahl WA, Zheng CY, Qi SR, Jin LY, Zhang CM, Gu LK, He JQ, Deng DJ, Ambudkar IS, and Wang SL. Sialin (SLC17A5) functions as a nitrate transporter in the plasma membrane. Proc Natl Acad Sci U S A 109: 13434–13439, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu JA, Wilson HL, and Rajagopalan KV. Structure-based alteration of substrate specificity and catalytic activity of sulfite oxidase from sulfite oxidation to nitrate reduction. Biochemistry 51: 1134–1147, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raat NJ, Noguchi AC, Liu VB, Raghavachari N, Liu DL, Xu XL, Shiva S, Munson PJ, and Gladwin MT. Dietary nitrate and nitrite modulate blood and organ nitrite and the cellular ischemic stress response. Free Radic Biol Med 47: 510–517, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajapakshe A, Meyers KT, Berry RE, Tollin G, and Enemark JH. Intramolecular electron transfer in sulfite-oxidizing enzymes: probing the role of aromatic amino acids. J Biol Inorg Chem 17: 345–352, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Laufs T, Reuss S, Hankeln T, and Burmester T. Divergent distribution of cytoglobin and neuroglobin in the murine eye. Neurosci Lett 374: 207–211, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Shiva S, Wang X, Ringwood LA, Xu X, Yuditskaya S, Annavajjhala V, Miyajima H, Hogg N, Harris ZL, and Gladwin MT. Ceruloplasmin is a NO oxidase and nitrite synthase that determines endocrine NO homeostasis. Nat Chem Biol 2: 486–493, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Shiva S, Huang Z, Grubina R, Sun JH, Ringwood LA, MacArthur PH, Xu XL, Murphy E, Darley-Usmar VM, and Gladwin MT. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res 100: 654–661, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Sparacino-Watkins CE, Tejero J, Sun B, Gauthier MC, Thomas J, Ragireddy V, Merchant BA, Wang J, Azarov I, Basu P, and Gladwin MT. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J Biol Chem 289: 10345–10358, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srihirum S, Sriwantana T, Unchern S, Kittikool D, Noulsri E, Pattanapanyasat K, Fucharoen S, Piknova B, Schechter AN, and Sibmooh N. Platelet inhibition by nitrite is dependent on erythrocytes and deoxygenation. PLoS One 7: e30380, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolz JF. and Basu P. Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3: 198–206, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Sturms R, DiSpirito AA, and Hargrove MS. Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry 50: 3873–3878, 2011 [DOI] [PubMed] [Google Scholar]
- 45.Tejero J. and Gladwin MT. The globin superfamily: functions in nitric oxide formation and decay. Biol Chem 395: 631–639, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temple CA, Graf TN, and Rajagopalan KV. Optimization of expression of human sulfite oxidase and its molybdenum domain. Arch Biochem Biophys 383: 281–287, 2000 [DOI] [PubMed] [Google Scholar]
- 47.Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, Ho C, Shiva S, Kim-Shapiro DB, and Gladwin MT. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem 286: 18277–18289, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Totzeck M, Hendgen-Cotta UB, Luedike P, Berenbrink M, Klare JP, Steinhoff HJ, Semmler D, Shiva S, Williams D, Kipar A, Gladwin MT, Schrader J, Kelm M, Cossins AR, and Rassaf T. Nitrite regulates hypoxic vasodilation via myoglobin-dependent nitric oxide generation. Circulation 126: 325–334, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utesch T. and Mroginski MA. Three-dimensional structural model of chicken liver sulfite oxidase in its activated form. J Phys Chem Lett 1: 2159–2164, 2010 [Google Scholar]
- 50.Veldman A, Santamaria-Araujo JA, Sollazzo S, Pitt J, Gianello R, Yaplito-Lee J, Wong F, Ramsden CA, Reiss J, Cook I, Fairweather J, and Schwarz G. Successful treatment of molybdenum cofactor deficiency type A with cPMP. Pediatrics 125: E1249–E1254, 2010 [DOI] [PubMed] [Google Scholar]
- 51.Webb A, Bond R, McLean P, Uppal R, Benjamin N, and Ahluwalia A. Reduction of nitrite to nitric oxide during ischemia protects against myocardial ischemia-reperfusion damage. Proc Natl Acad Sci U S A 101: 13683–13688, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb AJ, Patel N, Loukogeorgakis S, Okorie M, Aboud Z, Misra S, Rashid R, Miall P, Deanfield J, Benjamin N, MacAllister R, Hobbs AJ, and Ahluwalia A. Acute blood pressure lowering, vasoprotective, and antiplatelet properties of dietary nitrate via bioconversion to nitrite. Hypertension 51: 784–790, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamasaki H. and Sakihama Y. Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468: 89–92, 2000 [DOI] [PubMed] [Google Scholar]
- 54.Zuckerbraun BS, Shiva S, Ifedigbo E, Mathier MA, Mollen KP, Rao J, Bauer PM, Choi JJ, Curtis E, Choi AM, and Gladwin MT. Nitrite potently inhibits hypoxic and inflammatory pulmonary arterial hypertension and smooth muscle proliferation via xanthine oxidoreductase–dependent nitric oxide generation. Circulation 121: 98–109, 2010 [DOI] [PubMed] [Google Scholar]
- 55.Zuckerbraun BS, George P, and Gladwin MT. Nitrite in pulmonary arterial hypertension: therapeutic avenues in the setting of dysregulated arginine/nitric oxide synthase signalling. Cardiovasc Res 89: 542–552, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.