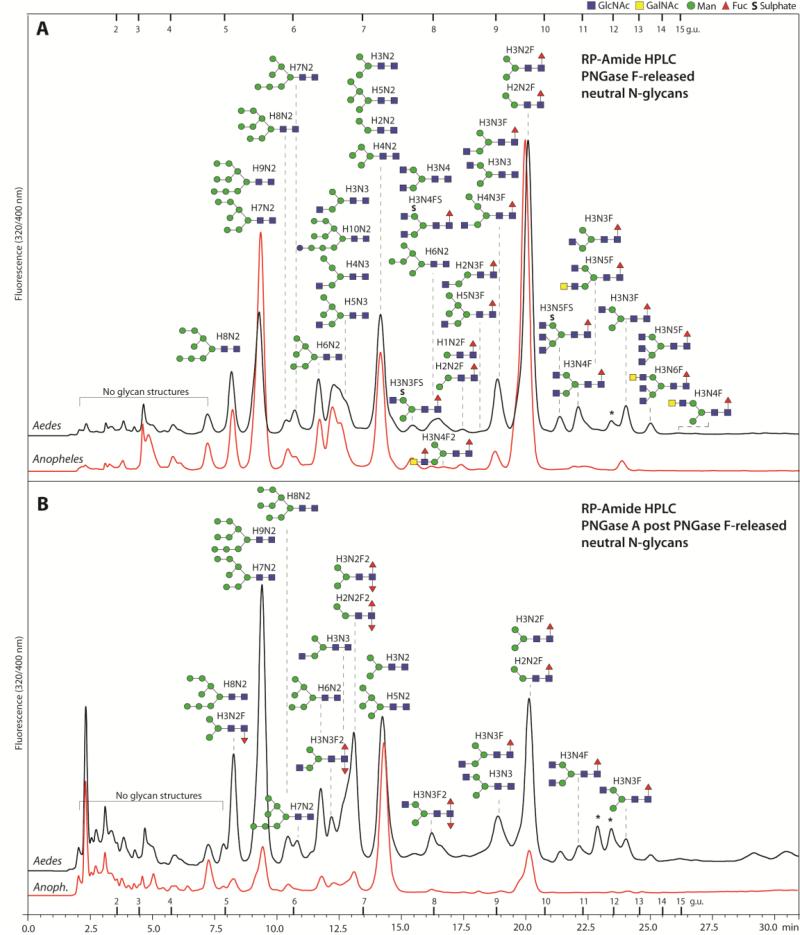
Figure 3. RP-Amide HPLC of mosquito neutral N-glycans.
The pyridylaminated neutral N-glycan pools of both mosquito species (Aedes and Anopheles) released by PNGase F (A) and PNGase A post PNGase F (B) were fractionated using a RP-Amide column. Selected peaks are annotated with the proposed N-glycan structures using the symbolic nomenclature of the Consortium for Functional Glycomics (circles, hexose; squares, N-acetylhexosamine; triangles, deoxyhexose; S, sulphate) as well as abbreviated compositions of the form HxNyFz; glycans eluting in one peak are shown according to relative intensity in the MALDI-TOF MS spectra (most abundant uppermost). The downward or upward depiction of the core fucose indicates respectively an α1,3 or α1,6-linkage; most of the monofucosylated species in the PNGase A post F chromatogram are α1,6-fucosylated as judged by retention time, whereas the occurrence of GalNAc is presumed from other studies on insect N-glycans. The elution positions of dextran standards are annotated with the relevant glucose units and peaks representing structures derived from the larval food source are marked with asterisks. The chromatograms are normalised with respect to each other; the original scales are: 4000 and 1850 mV (A. gambiae and A. aegypti F digest) and 125 and 85 (A. gambiae and A. aegypti A after F digest).