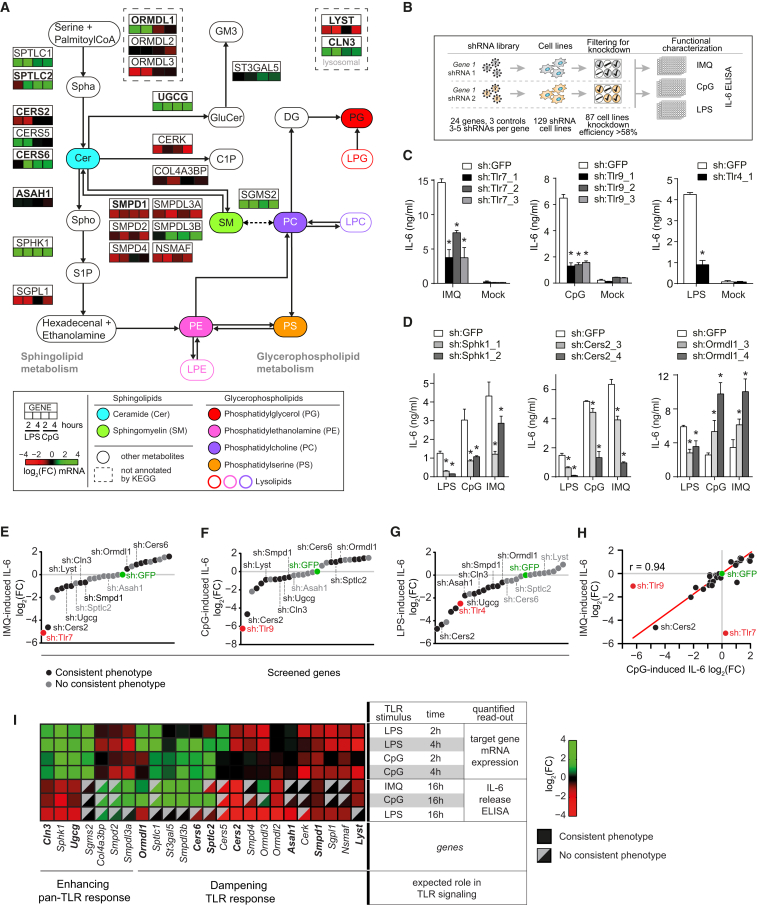
Figure 1.
TLR-Driven Transcription of the Sphingolipid Metabolic Network and Characterization of Cytokine Release upon shRNA-Mediated Silencing of This Network
(A) Selected sphingolipid and glycerophospholipid metabolic reactions (KEGG), shown together with main metabolites (rounded rectangles) and 24 selected proteins (rectangles). Protein location based on KEGG where possible. Heatmaps show relative expression of 24 selected genes after stimulation of RAW cells with LPS (100 ng/ml) or CpG (5 μM) for indicated time points measured by qRT-PCR. Bold protein names indicate selection for lipidomics analysis. Metabolites are colored consistently throughout the study. Data are combined of at least two independent experiments with technical triplicates. FC, fold-change; Spha, sphinganine; Spho, sphingosine; C1P, ceramide-1-phosphate. For other abbreviations, see text or legend and Table S1.
(B) Schematic representation of the generation and characterization of stable shRNA RAW cell lines, filtered based on knockdown efficiency.
(C) IL-6 release as measured by ELISA in sh:Tlr and sh:GFP control cell lines stimulated with IMQ (5 μM), or LPS (100 ng/ml) or CpG (5 μM) for 16 hr. Data are representative of at least five independent experiments and shown as mean ± SD of four technical replicates. ∗p < 0.0001.
(D) As in (C), but for sh:Sphk1, sh:Cers2, and sh:Ormdl1 cell lines. Data are representative of at least five independent experiments and shown as mean ± SD of four technical replicates. ∗p < 0.005.
(E–G) Screening results of three IL-6 release screens in 87 loss-of-function cell lines stimulated for 16 hr with IMQ, CpG, and LPS as measured by ELISA. Values are plotted as log2 fold-change relative to the respective sh:GFP control cell line and averaged over multiple shRNA cell lines. Black dots represent the averages of two or more shRNA cell lines with consistent phenotypes, while gray dots represent averages of all shRNA cell lines per gene. Indicated genes are selected for lipidomics analysis. Data are combined of at least five independent experiments.
(H) Scatter plot of IMQ and CpG screening results. Red line indicates linear fit. Data are combined of at least five independent experiments.
(I) Heatmap shows integration of target gene expression in wild-type RAW cells after stimulation with LPS and CpG and IL-6 release screening results of shRNA cell lines. Gray triangles indicate absence of consistent phenotypes for multiple shRNAs per gene. Data are combined of at least five independent experiments.