Figure 2.
Quantitative Lipidomics of Nine Stable shRNA Cell Lines Targeting Sphingolipid Metabolism Reveals Strongly Altered Lipid States
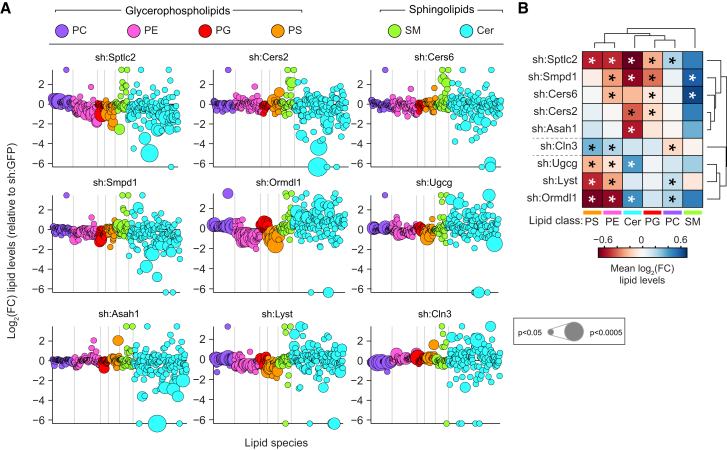
(A) Lipidomics analysis of nine loss-of-function cell lines. Values are shown as log2 fold-change relative to sh:GFP. Each dot represents a lipid species, color coded per lipid class. Dot size indicates significance. Lipidomics data are combined of three independent experiments and represented as mean.
(B) Hierarchical clustering of average log2 fold-change lipid levels per lipid class and cell line. Lipidomics data are combined of three independent experiments and represented as mean. ∗p < 0.05.