Figure S1.
TLR-Driven Transcription of the Sphingolipid Metabolic Network and Characterization of Cytokine Release upon shRNA-Mediated Silencing of This Network, Related to Figure 1
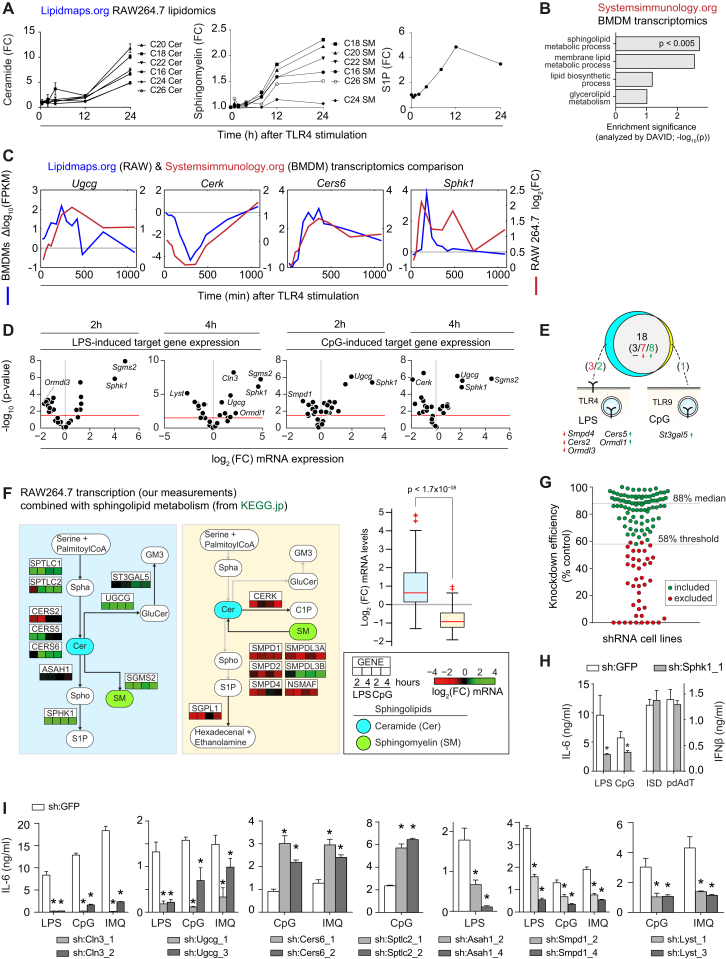
(A) TLR4-induced changes in the abundance of selected sphingolipids in RAW macrophages. Data from http://lipidmaps.org (Dennis et al., 2010).
(B) Pathway enrichment analysis of all differentially regulated genes in a genome-wide analysis of TLR4-stimulated bone marrow-derived macrophages (BMDMs). Shown are the highest enriched lipid-related annotations. Data from http://systemsimmunology.org (Ramsey et al., 2008). Enrichment analyzed by DAVID.
(C) Relative expression of key regulators of sphingolipid metabolism upon TLR4 stimulation over indicated time points in BMDMs and RAW cells. Relative expression calculated as delta log10 of the FPKM, or as log2 fold-change.
(D) Scatter plot of log2 fold-change expression (x axis) versus significance (y axis; t test) of RAW macrophages stimulated with LPS or CpG for 2 and 4 hr. Red lines indicate p < 0.05. Strongest regulated genes are indicated. Data are combined of two independent experiments with two technical replicates each.
(E) Venn diagram shows the number of regulated genes upon TLR4 stimulation by LPS and/or TLR9 stimulation by CpG. Predominant TLR localizations indicated in schemas. Red and green numbers in brackets indicate down- and upregulated genes respectively.
(F) LPS- and CpG-induced relative expression of selected genes separated by different branches of the sphingolipid metabolic pathway (KEGG). Boxplots group all expression values per subnetworks, with colors corresponding to subnetwork background colors. Abbreviations are as in Figure 1. Data are combined of two independent experiments with two technical replicates each.
(G) Knockdown efficiency measured by qRT-PCR of all 129 shRNA cell lines, normalized to sh:GFP. Each dot represents one cell line. Green dots were included in the screen, red dots were excluded due to insufficient knockdown efficiency. Threshold and median knockdown efficiency are indicated. Data are mean of technical triplicates.
(H) IL-6 release after stimulation with LPS or CpG, and IFNβ release after stimulation with Interferon-stimulatory DNA (ISD) or pdAdt in sh:Sphk1_1 and sh:GFP. Data are representative of three independent experiments.
(I) IL-6 release after stimulation with LPS, CpG or IMQ after 16 hr measured in selected shRNA cell lines and sh:GFP. ∗ indicates p < 0.005. Data are representative of five independent experiments and shown as mean ± SD of four technical replicates.
See also Table S1.