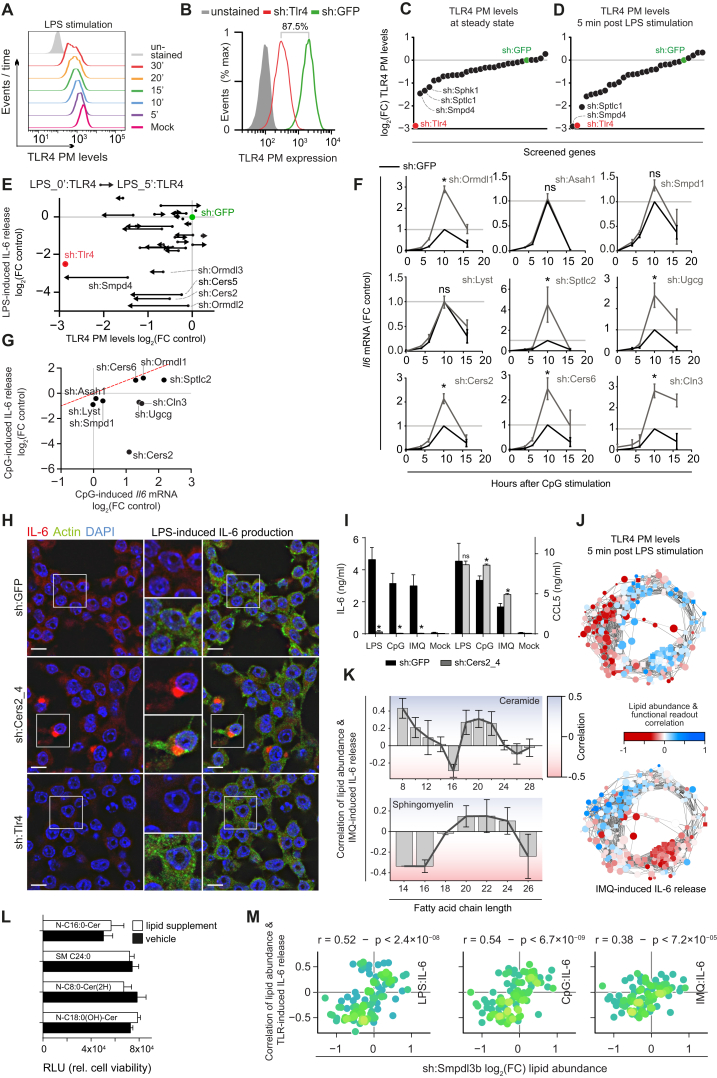
Figure S4.
Inference and Validation of Lipid Function in TLR-Related Processes, Related to Figure 4
(A) Histograms of TLR4-PE PM levels measured by FACS at steady state or after LPS (100ng/ml) stimulation at indicated time points in wild-type RAW cells.
(B) Histogram of steady-state TLR4-PE PM levels measured in sh:Tlr4 and sh:GFP cell lines analyzed by FACS.
(C and D) Screening results of TLR4 PM levels unstimulated (C) and after 5 min (D) of LPS (100 ng/ml) stimulation in loss-of-function cell lines stained with TLR4-PE and measured by FACS. Values are log2 fold-change of mean fluorescence intensity relative to sh:GFP. Indicated are genes with strongest knockdown phenotypes.
(E) Vector plot of log2 fold-change TLR4 PM levels from 0 to 5 min (x axis) versus log2 fold-change in LPS-induced IL-6 release (y axis). Vector origin (dot) indicates 0 min and end (arrow) indicates 5 min.
(F) Time course measurements of CpG-induced Il6 transcription in the nine knockdown cell lines used for lipidomics (gray line) normalized to unstimulated and 10h sh:GFP control (black line).
(G) Scatter plot of log2 fold-change CpG-induced Il6 mRNA levels (x axis) versus log2 fold-change in CpG-induced IL-6 release (y axis). Indicated are the nine genes selected for lipidomics analysis.
(H) Immunofluorescence microscopy of IL-6 protein levels in sh:Cers2_4, sh:GFP and sh:Tlr4 reveals perinuclear accumulation after 8h stimulation with LPS in sh:Cers2_4. IL-6 (red), actin (green), DAPI (blue). Scale bars indicate 10μm. Inserts show close-ups of indicated areas.
(I) IL-6 and CCL5 release after stimulation with LPS, CpG, or IMQ, in sh:GFP and sh:Cers2_4.
(J) Correlations between relative lipid abundance and measurements of LPS-induced TLR4 PM levels (top) and IMQ-induced IL-6 release (bottom) plotted on the circular network. Nodes of the network are color coded based on the strength of the correlation as indicated in legend.
(K) Average (gray bars) and SEM of the correlations between lipid abundance and IMQ-stimulated IL-6 release, per lipid fatty acid chain length, for ceramides (top) and sphingomyelins (bottom). Dark gray lines indicate chain length trends. Background colors vary with strength of correlation (red for negative, blue for positive correlations).
(L) Cell viability as measured by CellTiter-Glo luminescence, expressed in relative luminescence units (RLU) after supplementation with selected lipids (gray) or respective vehicle control (black).
(M) Scatter plots between relative lipid abundance independently measured for sh:Smpdl3b (x axis) against functional lipid correlations (y axis) for all measured TLR-induced IL-6 release. Dots represent individual lipids, colored based on the local data density. Strong and significant positive correlations of IL-6 release predict a pro-inflammatory phenotype, as confirmed (Heinz et al., 2015).
P-values are indicated above panels. (A) and (B) Data are representative of at least two independent experiments. (C) and (D) Data are combined of two independent experiments with two technical replicates each. (F) Transcriptional data are combined of two independent experiments and shown as mean ± SEM (H) Microscopy results are representative of two independent experiments. (I) Data are representative of at least two independent experiments. ∗ indicate p < 0.05. ns: not significant. (L) Data are representative of at least three independent experiments.