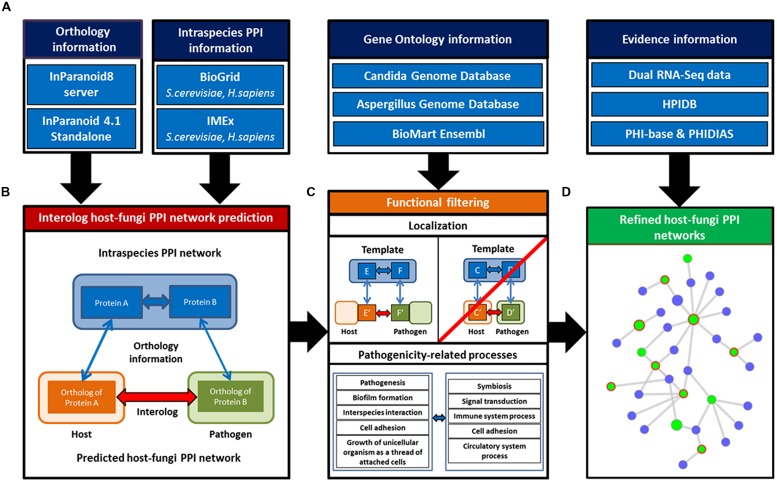
FIGURE 1.
Basic concept of the host–fungi PPI inference and refinement steps. (A) Information of direct PPI from multiple public databases were integrated for the two template networks Homo sapiens and Saccharomyces cerevisiae. (B) These combined with orthology information allowed to identify host–fungi interologs. (C) Primary inferred networks were filtered for interactions which showed protein localizations pointing to possible interfaces between host and fungi. Additionally, the networks were refined for pathogenicity-related processes. (D) Evidence information of several independent sources (e.g., transcriptome data) were exploited to evaluate the refined host–fungi PPI networks.