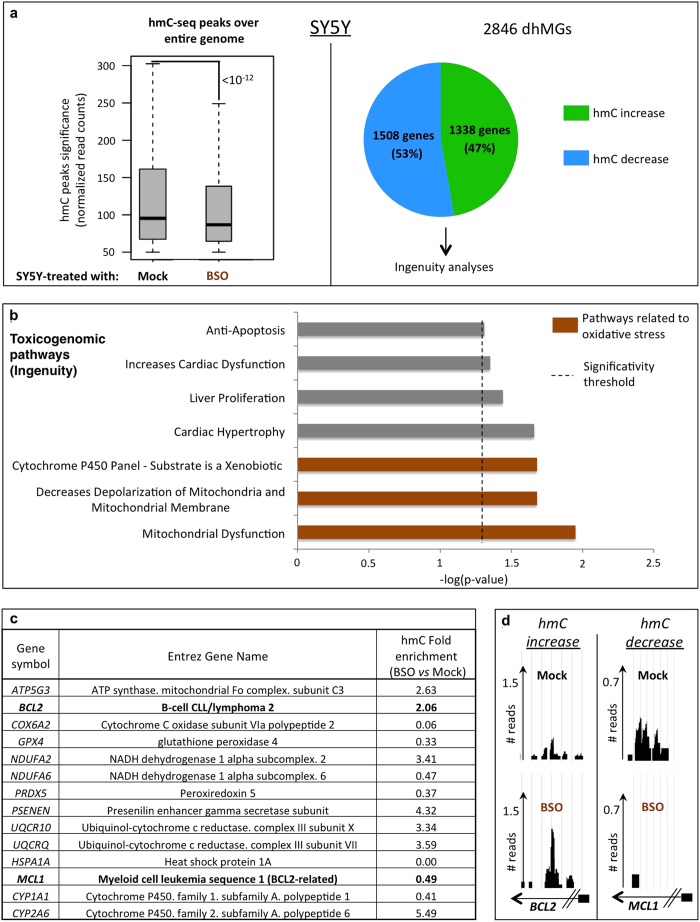
Figure 2. Genome-wide hmC sequencing after cell treatment with BSO highlights oxidative-stress-related pathways.
(A) Illumina deep sequencing was done on DNA from mock- and BSO-treated SY5Y cells after selective isolation of hydroxymethylated DNA fragments. Upper left panel: The global decrease in hmC (already demonstrated on dot blot and mass spectrometry, see Fig. 1B) is illustrated by a significant decrease (p-value < 10−12) in the normalized hmC- read count in BSO-treated cells. Upper right panel: Differentially hydroxymethylated genes (dhMGs) used in Ingenuity analyses. (B) Ingenuity toxicogenomic pathway analysis (“IPA-Tox”) of the dhMGs identified in SY5Y cells shows over-representation of oxidative-stress-related pathways (marked in brown). The x axis represents the log(p-value) and the dashed line shows the significance threshold for pathway over-representation. (C,D) Gene list and UCSC sequencing tracks associated with the oxidative-stress-related pathways shown in Fig. 2B. Gene symbols, Entrez gene names, and hmC fold changes for BSO- vs. mock-treated SY5Y cells are depicted. Normalized BCL2 and MCL1 read counts are represented in (D) as examples. Small black boxes attached to arrows represent the promoters of these genes. See also supplementary Fig. S2.