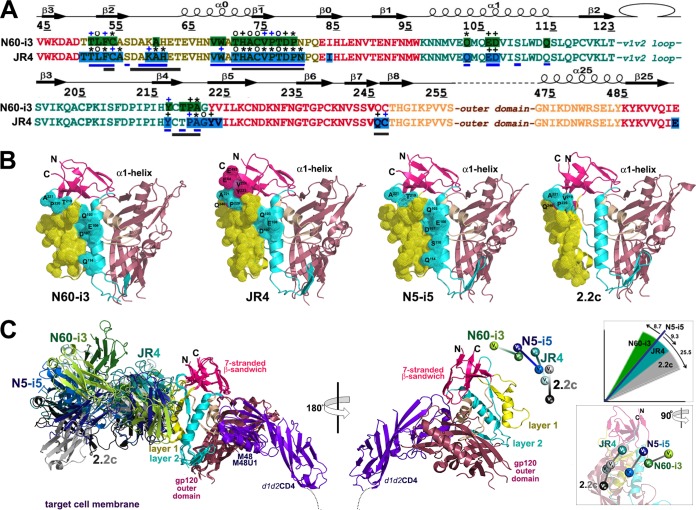
FIG 6.
Structural basis for interaction of cluster A MAbs with gp120 antigen. (A) Mapping of the N60-i3 and JR4 contact residues on the primary sequence of the gp120 inner domain of the 93TH057 isolate. The topology diagram depicting a distribution of secondary structure elements is shown above the gp120 sequences. Buried residues are highlighted in green and blue. Main chain (o), side chain (+), and both side and main chain (*) interactions are shown immediately above the residues as defined by a 5-Å distance criterion cutoff and colored based on contact type: hydrophobic, blue; hydrophilic, green; or both, black. Residues forming the N5-i5 and 2.2c epitopes as described in reference 53 are indicated by blue and gray lines below the gp120 sequence, respectively. (B) Epitope footprints of MAb N60-i3, JR4, N5-i5, and 2.2c. The Cα atoms of the gp120 residues involved in Fab binding are shown as spheres and displayed over the gp120 ribbon diagram. The selected residues of layer 2 and of the seven-stranded β-sandwich contribution to Fab binding and all the residues in the α1-helix involved in N60-i3, JR4, and N5-i5 binding are labeled. (C) Comparison of binding of MAb N60-i3, JR4, N5-i5, and 2.2c to CD4-triggered gp120 antigen. The N60-i3 Fab-gp12093TH057 coree-M48, JR4 Fab-gp12093TH057 coree-M48U1, and 2.2c Fab-gp120YU2 coree-M48U1 complexes were superimposed based on the gp120 outer domain onto the N5-i5 Fab-gp12093TH057 coree-d1d2CD4 complex and oriented relative to the target cell membrane. In the 180° view only the gp12093TH057 coree and d1d2CD4 from N5-i5 Fab-gp12093TH057 coree-d1d2CD4 complex (53) are shown, with variable heavy and light (VH and VL) domains of Fabs displayed as balls. Insets show rotation angles calculated using gp120's center of mass as an origin and the average α-carbon position for the heavy chain framework region 2 (residues 36 to 49) as a reference point for each antibody (top) and a 90°rotation of the 180°view (bottom).