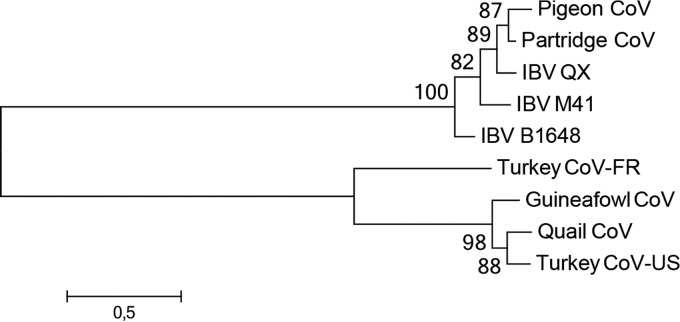
FIG 1.
Phylogenetic analysis of the S1 proteins of selected avian gammacoronaviruses. Amino acid sequences were aligned with ClustalX under default settings. The unrooted maximum likelihood tree was constructed by using MEGA6.06 with the best-fit substitution model with complete deletion (WAG+G+F model) and 100 bootstrap replicates (values indicated at the nodes). Pigeon CoV, pigeon coronavirus strain PSH050513 (GenBank accession number AAZ85066.1); Partridge CoV, partridge coronavirus strain GD/S14/2003 (accession number AAT70772.1); IBV QX, IBV strain QX (accession number AAF19629.1); IBV M41, IBV strain Mass 41 (accession number AAW33786.1); IBV B1648, IBV Belgian isolate B1648 (accession number CAA60684.1); Turkey CoV-FR, turkey coronavirus isolate FR070341j from France (accession number ADK88938.1); Guineafowl CoV, guineafowl coronavirus strain GfCoV/FR/2011 (accession number CCN27367.1); Quail CoV, quail coronavirus isolate quail/Italy/Elvia/2005 (accession number ABO45243.1); Turkey CoV-US, turkey coronavirus isolate ATCC from the United States (accession number ABW75138.1).