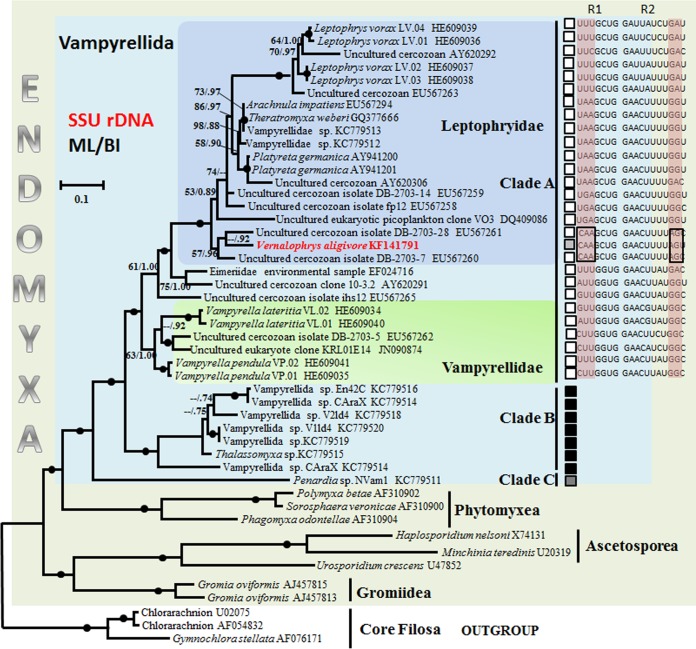
FIG 8.
SSU rDNA phylogeny of vampyrellids, showing the position of V. algivore with other published vampyrellid isolates and related environmental clones from GenBank. Three chlorarachniophytes were used to root the tree. This is the maximum-likelihood tree obtained by PhyML analyses (34) of 47 sequences using 2,148 aligned characters. Bootstrap support values after 1,000 replicates and Bayesian posterior probabilities are indicated at nodes when they are above 50% and 0.70, respectively. The black circles represent support values at or above 90%/0.95. Vampyrellids cluster in three main clades, A, B, and C. The black squares identify marine sequences, the white squares identify terrestrial (soil or freshwater) sequences, and the gray squares identify the species from brackish sediments. The aligned SSU rRNA sequences represent regions of helix E10_1 and E23_1. R1 and R2 represent partial sequences of E10_1 (nucleotides 117 to 123) and E23_1 (nucleotides 514 to 524), respectively, in the aligned SSU rRNA gene sequences; the nucleotide positions were based on the SSU rRNA gene sequence of Leptophrys vorax LV.03 (HE609038). Scale bar = 0.1, indicating that the sequence divergence is 10%.