ABSTRACT
Nitrate is widely used as a nitrogen source by cyanobacteria, in which the nitrate assimilation structural genes frequently constitute the so-called nirA operon. This operon contains the genes encoding nitrite reductase (nirA), a nitrate/nitrite transporter (frequently an ABC-type transporter; nrtABCD), and nitrate reductase (narB). In the model filamentous cyanobacterium Anabaena sp. strain PCC 7120, which can fix N2 in specialized cells termed heterocysts, the nirA operon is expressed at high levels only in media containing nitrate or nitrite and lacking ammonium, a preferred nitrogen source. Here we examined the genes downstream of the nirA operon in Anabaena and found that a small open reading frame of unknown function, alr0613, can be cotranscribed with the operon. The next gene in the genome, alr0614 (narM), showed an expression pattern similar to that of the nirA operon, implying correlated expression of narM and the operon. A mutant of narM with an insertion mutation failed to produce nitrate reductase activity, consistent with the idea that NarM is required for the maturation of NarB. Both narM and narB mutants were impaired in the nitrate-dependent induction of the nirA operon, suggesting that nitrite is an inducer of the operon in Anabaena. It has previously been shown that the nitrite reductase protein NirA requires NirB, a protein likely involved in protein-protein interactions, to attain maximum activity. Bacterial two-hybrid analysis confirmed possible NirA-NirB and NarB-NarM interactions, suggesting that the development of both nitrite reductase and nitrate reductase activities in cyanobacteria involves physical interaction of the corresponding enzymes with their cognate partners, NirB and NarM, respectively.
IMPORTANCE Nitrate is an important source of nitrogen for many microorganisms that is utilized through the nitrate assimilation system, which includes nitrate/nitrite membrane transporters and the nitrate and nitrite reductases. Many cyanobacteria assimilate nitrate, but regulation of the nitrate assimilation system varies in different cyanobacterial groups. In the N2-fixing, heterocyst-forming cyanobacteria, the nirA operon, which includes the structural genes for the nitrate assimilation system, is expressed in the presence of nitrate or nitrite if ammonium is not available to the cells. Here we studied the genes required for production of an active nitrate reductase, providing information on the nitrate-dependent induction of the operon, and found evidence for possible protein-protein interactions in the maturation of nitrate reductase and nitrite reductase.
INTRODUCTION
Nitrate is widely used as a nitrogen source by plants, algae, fungi, and bacteria, and it represents an important inorganic nutrient for the incorporation of nitrogen into living matter. Nitrate assimilation involves nitrate uptake into the cells and reduction to ammonium (1), which is the form of inorganic nitrogen that is incorporated into carbon skeletons, with an important contribution being made by the glutamine synthetase-glutamate synthase (GS/GOGAT) cycle (2, 3). Whereas the nitrate uptake and reduction pathway is relatively well conserved among nitrate-assimilating organisms, the regulation of nitrate assimilation follows diverse schemes in different organisms (1). Cyanobacteria are oxygenic photoautotrophs that can generally use nitrate and ammonium as nitrogen sources, but ammonium is used in preference over nitrate (4, 5). Many cyanobacteria can also fix atmospheric N2, and some filamentous cyanobacteria carry out N2 fixation in specialized cells termed heterocysts, which are formed only when the filaments have no source of combined nitrogen available (6).
Reduction of nitrate to ammonium takes place in two successive steps catalyzed by nitrate reductase and nitrite reductase, respectively, which in cyanobacteria are ferredoxin-dependent enzymes (4). An operon or gene cluster including genes encoding nitrite reductase (nirA), a nitrate/nitrite uptake transporter, and nitrate reductase (narB) is found in numerous cyanobacteria (4, 5; see also cyanobacterial genomes at https://img.jgi.doe.gov/). In many freshwater cyanobacteria, including well-investigated organisms such as Synechococcus elongatus strain PCC 7942 (here referred to as S. elongatus) and Anabaena sp. strain PCC 7120 (here referred to as Anabaena), the nitrate assimilation nirA operon includes the nrtABCD genes encoding an ABC-type transporter for nitrate and nitrite (7, 8). Figure 1 shows a scheme of the Anabaena genomic region bearing the nirA operon. A number of genes, including nirB, ntcB, and cnaT, transcribed in the opposite orientation are found upstream of nirA. The nirB and ntcB genes are present in a similar position in the S. elongatus genome (4). Whereas ntcB and cnaT are required for production of nirA operon transcripts (see below), the nirB gene has been shown to be required for attaining maximum levels of nitrite reductase (9, 10). NirB is likely involved in protein-protein interactions and has been suggested to act as a scaffolding protein for maturation of nitrite reductase (9, 10). Two open reading frames (ORFs) conserved in numerous cyanobacteria are located downstream from narB in the nirA operon of Anabaena (Fig. 1). ORF alr0613 encodes a 155-amino-acid hypothetical protein with four possible transmembrane segments that shows some similarity to the phosphate starvation-inducible E (PsiE) protein from Escherichia coli, which is of unknown function. ORF alr0614, on the other hand, encodes a 153-amino-acid protein that exhibits 47% identity to the S. elongatus NarM protein, which is required for the proper development of nitrate reductase activity in this unicellular cyanobacterium (11).
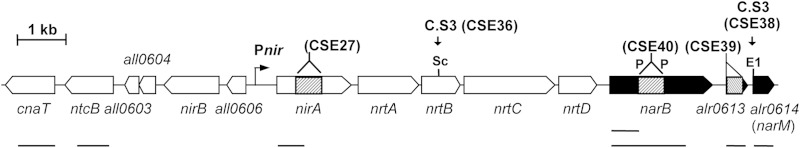
FIG 1.
Genomic region of Anabaena sp. strain PCC 7120 bearing the nitrate assimilation gene cluster. Genes and ORFs are indicated by thick arrows, which also show the direction of transcription. Black arrows correspond to the ORFs investigated in this work. The locations of the restriction sites into which gene cassette C.S3 (strains CSE36 and CSE38) was inserted are indicated. The regions deleted from nirA (strain CSE27; see reference 10), alr0613 (strain CSE39), and narB (strain CSE40) are indicated with hatched bars. Abbreviations for some restriction endonuclease sites: E1, EcoRI; P, PvuII; and Sc, ScaI. Lines below the genes denote the probes used for Northern analyses.
Nitrate reductase and nitrite reductase activities are generally lower in ammonium-grown than in nitrate-grown cyanobacterial cells (4). However, whereas in non-N2-fixing cyanobacteria, such as S. elongatus, expression of these enzymatic activities takes place at appreciable levels in the absence of nitrate or nitrite, in the heterocyst-forming, N2-fixing cyanobacteria, including Anabaena, induction by nitrate or nitrite is required for attaining high levels of expression in the absence of ammonium (4, 10, 12). Expression of the nirA operon upon ammonium withdrawal is promoted by NtcA, a cyclic AMP receptor protein-family transcription factor (13) whose activity is enhanced by 2-oxoglutarate (14–17) and PipX (18, 19). PipX is a small protein factor whose availability is controlled by the signal transduction PII protein, which sequesters PipX under low C-to-N balance conditions (18, 19). In addition to NtcA, a route-specific LysR-type transcriptional regulator, NtcB, is involved in the regulation of nirA operon expression (20–23). In contrast to NtcA, which is strictly required for expression of the nirA operon in all investigated cyanobacterial strains, NtcB is involved in regulation with different stringency levels, depending on the cyanobacterial strain. In the case of Anabaena, a strict requirement for NtcB for the expression of the nirA operon is observed (22). In S. elongatus, however, NtcB is not absolutely required for nitrate assimilation, although it mediates a nitrite-dependent enhancement of expression of the nirA operon in the presence of the glutamine synthetase inhibitor l-methionine-d,l-sulfoximine (20, 24). A third positive regulatory element of nirA operon expression in Anabaena is CnaT, which does not appear to be a DNA-binding protein and, consequently, may affect nirA operon expression indirectly (25). As is the case for NtcB, whereas CnaT is strictly required for nitrate assimilation in Anabaena, any phenotype related to nitrate assimilation was observed in an S. elongatus cnaT mutant (11). In addition to the positive elements described above, two elements negatively affecting the expression of the nirA operon have been identified in Anabaena. These are the nitrite reductase protein itself and NirB, which negatively regulate the expression of the nirA operon when nitrate is not available in the culture medium (10).
In this study, we aimed at characterizing the 3′ region of the nirA operon in Anabaena (Fig. 1). We found that, under certain culture conditions, alr0613 is coexpressed with the Anabaena nirA operon and that alr0614 is expressed monocistronically with a pattern similar to that of the nirA operon. Consistent with results previously described for S. elongatus (11), we found that alr0614 is required for production of nitrate reductase activity, corroborating that it is the Anabaena narM gene. Additionally, we isolated an Anabaena mutant with a deletion in the nitrate reductase structural gene narB (ORF alr0612). Studying the narM and narB mutants, we found that a functional nitrate reductase is required for attaining high levels of expression of the nirA operon in the presence of nitrate in the culture medium. Finally, possible interactions between nitrite reductase and NirB and between nitrate reductase and NarM were corroborated by a bacterial two-hybrid (BACTH) analysis.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena sp. (also known as Nostoc sp.) strain PCC 7120 was routinely grown photoautotrophically at 30°C under white light (about 25 μE s−1 m−2), with shaking being used for liquid cultures. The media used for growth were BG11 (with NaNO3 used as the nitrogen source [26]), BG110 (BG11 without nitrate), BG110NO2 (BG110 supplemented with 2 mM NaNO2), or BG110NH4+ [BG110 supplemented with 4 mM NH4Cl and 8 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)–NaOH buffer, pH 7.5]. For growth on plates, medium solidified with separately autoclaved 1% agar (Difco) was used. When appropriate, the following antibiotics were added to the plates at the indicated final concentrations: streptomycin sulfate (Sm) at 5 μg/ml and spectinomycin dihydrochloride pentahydrate (Sp) at 5 μg/ml. In liquid cultures, the antibiotic concentrations used were as follows: 2 μg/ml for Sm and 2 μg/ml for Sp. Strains CSE36 (nrtB) and CSE38 (alr0614) were routinely grown in BG110NH4+ medium supplemented with Sm and Sp. Strains CSE27 (nirA), CSE39 (alr0613), and CSE40 (narB) were routinely grown in BG110NH4+ medium. Derepression experiments were carried out as described previously (10).
Escherichia coli DH5α, HB101, XL1-Blue, and ED8654 were grown in Luria-Bertani medium as described previously (27). Strain BTH101 (cya-99) was used for BACTH analysis (28).
Generation of mutant strains.
The method of sacB-mediated positive selection for double recombinants in Anabaena (29) was used to generate mutant strains CSE36 (nrtB::C.S3), CSE38 (narM::C.S3), CSE39 (Δalr0613), and CSE40 (ΔnarB). Plasmids pCSE153B (for CSE36), pCSE140B (for CSE38), pCSE174B (for CSE39), and pCSE245 (for CSE40) were transferred to the cyanobacterial parental strain by conjugation (30). See Table 1 for descriptions of the strains and plasmids. Plasmids pRL623 and pRL443 were used as helper and conjugative plasmids in the conjugations, respectively (31). In all cases, the genomic structure of the resultant Anabaena mutant strain was checked by Southern analysis or PCR analysis to confirm the absence of wild-type chromosomes. All mutant strains were homozygous for the mutated chromosomes.
TABLE 1.
Cyanobacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| PCC 7120 | Wild-type Anabaena strain | 26 |
| EF116 | Derivative of Anabaena sp. strain PCC 7120 unable to fix nitrogen under oxic conditions | 47 |
| CSE27 | Derivative of EF116 with a deletion in the nirA gene | 10 |
| CSE36 | Smr Spr derivative of strain EF116; nrtB::C.S3 | This work |
| CSE38 | Smr Spr derivative of strain PCC 7120; narM::C.S3 | This work |
| CSE39 | Derivative of strain PCC 7120; Δalr0613 | This work |
| CSE40 | Derivative of strain PCC 7120; ΔnarB | This work |
| Plasmids | ||
| pCSE139 | 1,618-bp product of PCR with primers narB-7120-5 and alr0614-3, cloned in pGEM-T | This work |
| pCSE140B | pCSE139 insert bearing EcoRI-ended gene cassette C.S3 (39) inserted into the EcoRI site of narM, cloned in pRL271; used to generate mutant strain CSE38 | This work |
| pCSE148 | 2.0-kb BspHI/SphI fragment from pCSE26 (8) cloned in NcoI/SphI-digested pCSAM170 (48); it overproduces His-tagged Anabaena NirA protein | This work |
| pCSE153B | 2.9-kb ClaI/XbaI fragment from pCSE2 (8) bearing HincII-ended gene cassette C.S3 inserted into the ScaI site of nrtB, cloned in pRL278; used to generate mutant strain CSE36 | This work |
| pCSE174B | 1,265-bp product of PCR with primers narB-7120-5, alr0613-5, alr0613-6, and alr0614-3, with pCSE39 as the template; presents a deletion of a 351-bp internal segment of alr0613, corresponding to nucleotides 36 to 386 of the 459-nucleotide coding region; cloned in pRL277; used to generate mutant strain CSE39 | This work |
| pCSE189 | PCR fragment amplified using primers narB-7120-8 and narB-7120-11, cloned in pKT25 using XbaI and KpnI; it produces the T25-NarB hybrid protein | This work |
| pCSE191 | Derivative of pUT18C; it presents restriction site XbaI in a polylinker replaced by restriction site NcoI, using primers pUT18-3 and pUT18-4 | This work |
| pCSE192 | Derivative of pKT25; it presents restriction sites SalI and KpnI in a polylinker replaced by restriction sites BspHI and SphI, respectively, using primers pKT25-3 and pKT25-4 | This work |
| pCSE193 | 2.0-kb BspHI/EcoRV fragment from pCSE26 cloned in pCSE192 using BspHI and SmaI; it produces the T25-NirA hybrid protein | This work |
| pCSE195 | 1,716-bp product of PCR with primers orf398-7120-5 and all0604-2 (10), cloned in pCSE191 using NcoI and ClaI; it produces the T18-NirB hybrid protein | This work |
| pCSE200 | As for pCSE193, but cloned in pCSE191 using NcoI and SmaI; it produces the T18-NirA hybrid protein | This work |
| pCSE201 | Insert of pCSE195 cloned in pCSE192 using BspHI and SmaI; it produces the T25-NirB hybrid protein | This work |
| pCSE242 | PCR fragment amplified using primers alr0614-9 and alr0614-10, cloned in pUT18 using PstI and EcoRI; alr0614-9 also allows elimination of the EcoRI site internal to narM without changing the NarM amino acid sequence; it produces the T18-NarM hybrid protein | This work |
| pCSE243 | As for pCSE242, but cloned in pUT18C using PstI and EcoRI; it produces the NarM-T18 hybrid protein | This work |
| pCSE245 | narB gene with a deletion of the internal 588-bp PvuII fragment, cloned in pRL277; used to generate mutant strain CSE40 | This work |
| pCSE246 | As for pCSE189, but cloned in pKNT25 using XbaI and KpnI; it produces the NarB-T25 hybrid protein | This work |
| pRL277 | Smr Spr, sacB-carrying, mobilizable vector | 49 |
| pRL278 | Nmr, sacB-carrying, mobilizable vector | 49 |
| pRL443 | Kms derivative of conjugative plasmid RP4 | 30 |
| pRL623 | Mobilization helper; encodes M.AvaI, M.Eco47II, and M.EcoT22Ia | 31 |
| pUT18 | Plasmid used for BACTH analysis | 28 |
| pUT18C | Plasmid used for BACTH analysis | 28 |
| pKT25 | Plasmid used for BACTH analysis | 28 |
| pKNT25 | Plasmid used for BACTH analysis | 28 |
M, methylase.
RNA isolation and analysis.
RNA from the Anabaena spp. was prepared as described previously (32). The resulting RNA preparations were treated with RNase-free DNase I to eliminate contaminating DNA. For Northern blot analysis, RNA (approximately 20 to 25 μg) was subjected to electrophoresis in denaturing formaldehyde gels, transferred to Hybond-N+ membranes, and subjected to hybridization at 65°C as described previously (33). The DNA probes (see reference 10) used in Northern experiments were the nirA probe, a DNA fragment generated by PCR using primers nir-7120-23 and nir-7120-25; the narB probe, a DNA fragment generated by PCR using primers N-narB-7120 and C-narB-7120; the narB probe (5′ end), a 654-bp DNA fragment resulting from cutting with PvuII a DNA fragment generated by PCR using primers narB-7120-3 and narB-7120-4; the ntcB probe, a DNA fragment generated by PCR using primers Nc-ntcB and ntcB-3 (22); the cnaT probe, a HincII/BstXI DNA fragment from pCSE118 (25); the alr0613 probe, a DNA fragment generated by PCR using primers alr0613-1 and alr0613-2; and the alr0614 probe, a DNA fragment generated by PCR using primers alr0614-1 and alr0614-2. (The oligodeoxynucleotide primers used for the first time in this work are described in Table 2.) For PCR-generated probes, Anabaena genomic DNA was used as a template. Results were visualized and quantified with a Cyclone storage phosphor system and OptiQuant image analysis software (Packard).
TABLE 2.
Oligodeoxynucleotide primers used in this work
| Primer | Sequencea (5′–3′) |
|---|---|
| orf398-7120-5 | CCAAAGCCATGGCGAATAATATCGGTC |
| narB-7120-3 | CTACCAAAACCCTATGTCC |
| narB-7120-4 | GCGAATAGTGGCGAATTACC |
| narB-7120-5 | GCTCGAGCGTCGAGGTAGCGCCAAG |
| narB-7120-6 | CCACTGAGATGACTGGAG |
| narB-7120-8 | CGACGTTCTAGAATCTACCAAAACC |
| narB-7120-11 | AACAGGTACCCACTGAGATGACTGGAG |
| alr0613-1 | GGCAAATTTTGGGAATCACG |
| alr0613-2 | GACAGAGCTAAAATTGCCAC |
| alr0613-4 | CGTGATTCCCAAAATTTGCC |
| alr0613-5 | CACGGATCCGGGCGTGGCAATTTTAGCTC |
| alr0613-6 | GCCACGCCCGGATCCGTGATTCCCAAAATTTGCC |
| alr0614-1 | CAGATTTTGTTGATTCCCTGCG |
| alr0614-2 | AAACTCGGCGATCGCCTTGGG |
| alr0614-3 | ACTCGAGGATTGAGTTAGTAGTAAGGAC |
| alr0614-4 | GAATTCTGCCATAAATGAGCG |
| alr0614-9 | ATTCTGCAGTGAGTTCTTTGAATTTGAATCAG |
| alr0614-10 | ATTGAATTCCCTATTCTACTTATATTAAACTCG |
| pKT25-3 | GTCATGACTCTAGAGGATCCCGGGGCATGCCTAAGTAACTAAG |
| pKT25-4 | AATTCTTAGTTACTTAGGCATGCCCCGGGATCCTCTAGAGTCATGACTGCA |
| pUT18-3 | GGTCGACCATGGAGGATCCCCGGGTACCGAGCTCG |
| pUT18-4 | AATTCGAGCTCGGTACCCGGGGATCCTCCATGGTCGACCTGCA |
Introduced restriction enzyme-cutting sites are shown in boldface.
For reverse transcriptase PCR (RT-PCR) experiments, 10 μg of Anabaena total RNA was mixed, in a final volume of 20 μl, with 40 pmol of oligonucleotide narB-7120-6, alr0613-4, alr0613-2, alr0614-4, or alr0614-2 in the presence of 10 mM Tris-HCl (pH 8.0), 150 mM KCl, and 1 mM EDTA, and the mixture was heated for 10 min at 85°C and then at 50°C for 1 h for annealing. To control for the presence of contaminating DNA, 10 μl of an annealing reaction mixture was treated with 1 μg RNase A for 15 min at 50°C. The extension reactions were carried out at 47°C for 1 h in a final volume of 20 μl containing 10 μl of the annealing reaction mixture (with or without RNase), 0.5 mM each deoxynucleoside triphosphate, 100 U of reverse transcriptase (SuperScript II; Invitrogen), 10 mM dithiothreitol, and the buffer recommended by the transcriptase provider. PCR was carried out with 2 μl of a reverse transcription mixture as the template and the following oligonucleotide pairs as the primers: narB-7120-5/narB-7120-6 (for the segment named A in Fig. 3), narB-7120-5/alr0613-4 (for segment B), narB-7120-5/alr0613-2 (for segment C), narB-7120-5/alr0614-4 (for segment D), narB-7120-5/alr0614-2 (for segment E), alr0613-1/alr0613-2 (for segment F), alr0613-1/alr0614-4 (for segment G), alr0613-1/alr0614-2 (for segment H), and alr0614-1/alr0614-2 (for segment I). PCRs with samples containing the same oligonucleotides and plasmid pCSE139 as the template were run in parallel and used as controls. PCR was performed by standard procedures, and the PCR products were resolved by electrophoresis in 0.7% agarose gels.
FIG 3.
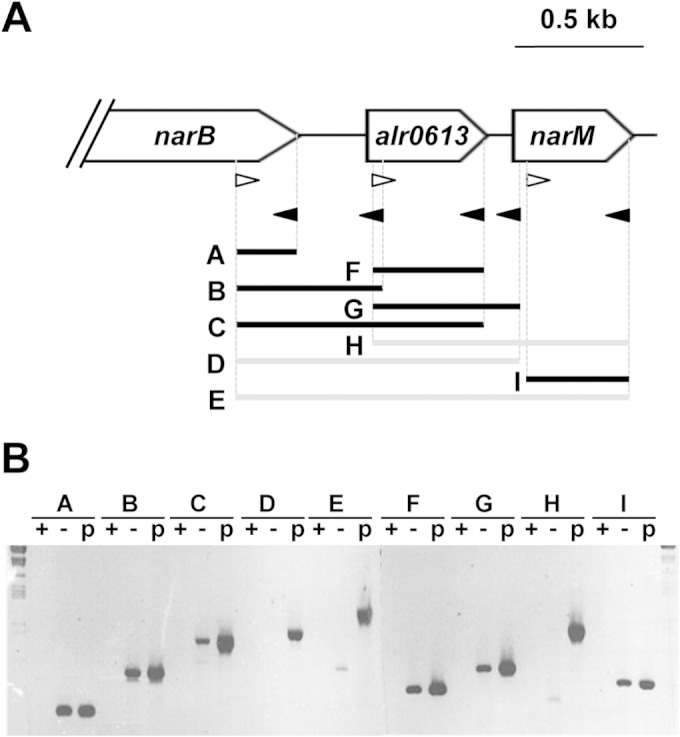
RT-PCR analysis of expression of the narB to narM (alr0612 to alr0614) region. (A) The gene region and Anabaena gene/ORF names (46) are depicted. (B) Reverse transcription was carried out with the oligonucleotide primers depicted as closed arrowheads in panel A and with RNA isolated from cells grown with ammonium and incubated for 4 h in medium containing nitrate. (Open arrowheads in panel A represent forward primers used for amplification by PCR.) The positions of the primers used for amplification correspond to the ends of the segments indicated in panel A, which are depicted in black, indicating amplification by RT-PCR, or in gray, indicating no amplification. Lanes + and −, RNA samples treated and RNA samples not treated with RNase A, respectively; lanes p, plasmid pCSE139 used as the template for amplification with the corresponding primers.
BACTH assays.
The plasmids used for the BATCH assays (28) were cotransformed into E. coli BTH101 (cya-99). Four transformants of each combination of plasmids were plated onto LB medium containing selective antibiotics, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg ml−1), and 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and then incubated at 30°C for 24 to 48 h. The efficiencies of the interactions between different hybrid proteins were quantified by measuring β-galactosidase activity in liquid cultures. Bacteria were grown in LB medium in the presence of 0.5 mM IPTG and appropriate antibiotics at 30°C for 16 h. Before the assays, the cultures were diluted 1:5 into buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4). To permeabilize cells, 30 μl of toluene and 35 μl of a 0.1% SDS solution were added to 2.5 ml of bacterial suspension. The tubes were vortexed for 10 s and incubated with agitation at 37°C for 45 min for evaporation of toluene. For the enzymatic reaction, 875 μl of permeabilized cells was added to buffer Z supplemented with β-mercaptoethanol (final concentration, 25 mM) to a final volume of 3.375 ml. The tubes were incubated at 30°C in a water bath for at least 5 min. The reaction was started by adding 875 μl of 0.4 mg ml−1 o-nitrophenol-β-galactoside in buffer Z without β-mercaptoethanol. One-milliliter samples, taken at times for up to 10 min, were added to 0.5 ml of 1 M Na2CO3 to stop the reaction. The A420 was recorded, and the amount of o-nitrophenol (ONP) produced was calculated using an extinction coefficient (ε420) of 4.5 mM−1 cm−1 and referred to the amount of total protein, determined by a modified Lowry procedure (34). The amount of ONP produced per milligram of protein versus time was plotted and β-galactosidase activity was deduced from the slope of the linear function.
Expression and purification of Anabaena NirA and production of antibodies.
Plasmid pCSE148, which contains the Anabaena nirA gene fused to a sequence encoding a 6-His tag under an IPTG-inducible promoter (Table 1), was transferred to E. coli XL1-Blue. An 80-ml preinoculum of this strain grown overnight in LB medium with 50 μg of ampicillin (Ap) ml−1 and 2% glucose was used to inoculate 720 ml of LB medium with 50 μg Ap ml−1 and 6 mg liter−1 of ferric ammonium citrate. The culture was incubated at 37°C for 1 h with shaking. Protein expression was induced by addition of 0.5 mM IPTG. After 3 h at 37°C, cells were collected, washed in buffer A (40 mM Tris-HCl, pH 7.5, 300 mM NaCl), and resuspended in buffer A (5 ml/g of cells) with 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were broken by passage twice through a French pressure cell at 14,000 lb/in2. After centrifugation at 8,000 × g (15 min, 4°C), the insoluble 6His-NirA protein was purified by solubilization of inclusion bodies in a buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 8 M urea (35) and chromatography of solubilized proteins through a 5-ml His-Select column from Sigma, using buffer A with 6 M urea and imidazole to elute the retained proteins. Samples obtained after purification (50 to 65 mM imidazole) were subjected to SDS-PAGE, excised from the gel, electroeluted, and concentrated (Amicon Ultra 4; Millipore). An amount of 1.0 mg of purified protein was used for the subcutaneous injection of a rabbit to produce antibodies in the Centro de Producción y Experimentación Animal, Universidad de Sevilla (Seville, Spain). These experiments were performed in accordance with the European Union Directive 2010/63/EU on the protection of animals used for scientific purposes and approved by the Committee of Animal Use for Research at the Universidad de Sevilla. Antiserum was recovered 65 days after the first injection and stored at −80°C until use.
Anabaena cell extracts and immunoblotting.
For obtaining cell extracts, filaments (corresponding to about 60 μg of chlorophyll a) from strains PCC 7120, CSE38, and CSE40 grown in ammonium (time zero [t0]) and incubated in medium containing ammonium, nitrate, nitrite, or no combined nitrogen for 4 h were harvested by filtration, washed with a buffer containing 50 mM Tris-HCl and 100 mM EDTA (pH 8), and, finally, resuspended in a buffer containing 10 mM HEPES-NaOH (pH 8), 5 mM MgCl2, 2.5 mM CaCl2, 10% glycerol, and 1 mM PMSF. The filaments were broken, after adding 50 μl of acid-treated glass beads (G9268; Sigma), by 10 cycles of vortexing (1 min) and keeping the samples in ice (1 min). Cell debris was discarded after centrifugation at 16,100 × g (20 min, 4°C), and the supernatant was stored at −20°C until use.
For immunoblotting, samples were loaded, run in a 10% Laemmli SDS-polyacrylamide gel, and transferred to polyvinylidene difluoride membrane filters. For detection of nitrite reductase (NirA), the filters were incubated overnight in blocking buffer containing 15 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5% nonfat milk powder, and 0.05% Nonidet P-40. Primary antibody (anti-NirA) was added at a dilution of 1:10,000 in buffer B (15 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2% nonfat milk powder, 0.25% Triton X-100) and incubated for 1 h at 30°C. The secondary antibody (anti-rabbit IgG conjugated to peroxidase [Sigma]) was added at a dilution 1:10,000 in buffer B. Detection was performed with a chemiluminescence kit (ECL plus; GE Healthcare) and exposure to Hyperfilm (GE Healthcare).
Enzyme activities and heterocyst counts.
Nitrate reductase (36) and nitrite reductase (37) were measured with dithionite-reduced methyl viologen as the reductant in cells made permeable with mixed alkyltrimethylammonium bromide. The amounts of the cells added to the enzymatic assays for nitrate reductase and nitrite reductase contained 5 and 25 μg of chlorophyll a, respectively. Activity units correspond to micromoles of nitrite produced (nitrate reductase) or removed (nitrite reductase) per minute.
To determine the percentage of heterocysts in filaments of Anabaena and mutant strains, samples of the cultures were examined by light microscopy, and a total of about 1,000 cells were counted for each strain and culture condition.
RESULTS
Expression of genes located downstream of narB.
ORFs alr0613 and alr0614 (narM) are located downstream of and in the same orientation as the narB gene in the genome of Anabaena (Fig. 1). Expression of alr0613 and narM was studied in wild-type Anabaena, a nirA mutant (strain CSE27) (10), and an nrtB mutant (strain CSE36). Strain CSE36 was constructed by insertion of gene cassette C.S3 into the nrtB gene (Fig. 1), which encodes the membrane component of the ABC-type nitrate/nitrite transport system in Anabaena (8). The C.S3 cassette bears transcription terminators in both ends (38, 39). Because of the polar effect that the insertion of C.S3 has on the expression of downstream genes, including narB in the nirA operon, strain CSE36 (nrtB) could not grow using nitrate as the nitrogen source. However, because nirA is upstream of the insertion site and because the transporter is not required in medium containing relatively high concentrations of nitrate or nitrite (8), strain CSE36 (nrtB) could grow using nitrite.
Gene expression was studied by Northern analysis, performed with probes for each of the indicated genes and RNA isolated from filaments grown with ammonium and incubated in the presence of ammonium, nitrate, or no combined nitrogen for 4 h (Fig. 2). As previously reported (8), in Anabaena the nirA operon produces a long transcript of a size close to 10 kb that is unstable, and therefore, only a smear of RNA molecules could be detected with the narB probe (Fig. 2A). In wild-type Anabaena, the alr0613 probe hybridized to a diffuse band of about 0.56 kb (likely corresponding to an alr0613 transcript) present under all conditions and to a faint smear of longer RNAs in nitrate-incubated cells (Fig. 2B). On the other hand, the narM probe hybridized to three bands of about 0.92, 0.77, and 0.62 kb, which were more abundant in the presence than in the absence of nitrate (Fig. 2C). In strain CSE27 (nirA), which overexpresses the nirA operon in the absence of ammonium (see the hybridization with the narB probe in Fig. 2A and reference 10), the smear detected with the alr0613 probe in nitrate-incubated wild-type cells was observed at a higher level both in the presence of nitrate and in the absence of combined nitrogen (Fig. 2B). Likewise, the accumulation of a higher level of the different narM transcripts was observed in strain CSE27 (nirA) than in the wild type in the presence of nitrate or in the absence of combined nitrogen (Fig. 2C). In contrast, in strain CSE36 (nrtB), in which no expression of genes downstream of nrtB takes place (see the expression of narB in Fig. 2A), neither the smear obtained with the alr0613 probe nor the high abundance of the different narM transcripts was observed in nitrate-incubated cells (Fig. 2B and C). These results indicate that the full expression of alr0613 and narM depends on completion of the nirA operon transcript. However, some hybridization signals were still observed with the alr0613 and narM probes in strain CSE36 (nrtB) under the different nitrogen regimes tested (Fig. 2B and C). These results may imply that both alr0613 and narM are cotranscribed with the nirA operon but also that they are expressed at a low level as independent transcripts.
FIG 2.
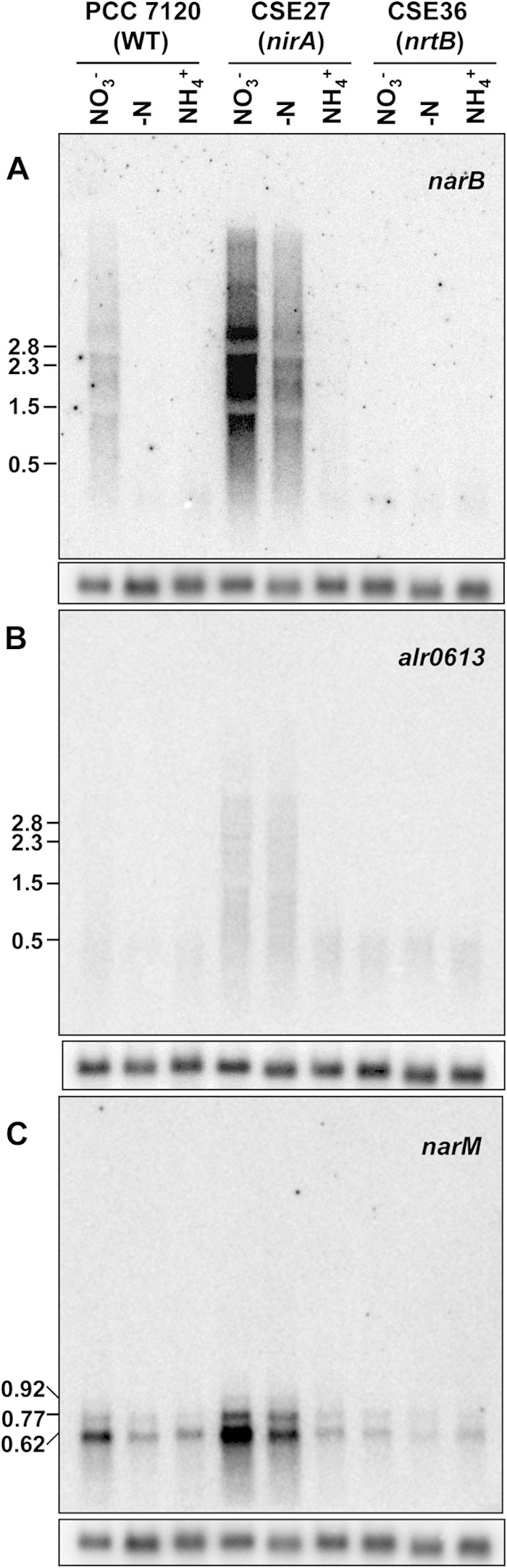
Expression of narB, alr0613, and narM (alr0614) in strains PCC 7120 (wild type [WT]), CSE27 (nirA), and CSE36 (nrtB). Hybridization assays were carried out using RNAs isolated from cells grown with ammonium and incubated for 4 h in medium containing nitrate (NO3−), ammonium (NH4+), or no combined nitrogen (−N). Hybridization to rnpB (45) served as a loading and transfer control for each of the filters used and is shown in the image below each panel. The positions of some size markers or identified transcripts (in kilobases) are shown on the left.
The expression of the narB to narM gene cluster depicted in Fig. 3A was also investigated by RT-PCR. To determine the transcriptional units in this genomic region, five distinct primers were used for reverse transcription and different primer pairs were used for PCR amplification. RNA was isolated from ammonium-grown wild-type cells incubated in nitrate-containing medium for 4 h. Detection of amplification products A, F, and I indicates that mRNA molecules corresponding to narB, alr0613, and narM, respectively, were present in the RNA preparation (Fig. 3B). Detection of amplification products B and C indicates the existence of transcripts overlapping narB and alr0613 (Fig. 3B). Additionally, detection of amplification product G but not that of amplification product D suggests that a distinct alr0613 transcript, likely more abundant than the part of the nirA operon transcript covering the same region (alr0613 down to the beginning of narM), was present in the RNA preparation. On the other hand, evidence for transcripts overlapping narB or alr0613 and the whole narM gene was not obtained (amplification product E or H, respectively, was not detected), although a distinct narM transcript was found (see amplification product I). Therefore, these results corroborate those obtained by Northern analysis, showing that there is a transcript covering the nirA operon and alr0613 in Anabaena cells incubated in the presence of nitrate and that individual transcripts of alr0613 and narM are produced as well. They do not support, however, the possibility that the whole narM gene can be cotranscribed with the nirA operon.
Isolation and characterization of alr0613, alr0614 (narM), and alr0612 (narB) mutants.
Given that alr0613 is coexpressed with the nirA operon under certain conditions and that narM presents an expression pattern similar to that of the nirA operon and encodes a protein likely related to nitrate assimilation, we inactivated both ORFs to test their possible role in nitrate assimilation in Anabaena. An alr0613 mutant, strain CSE39, was constructed by deletion of most of its coding sequence (see Fig. 1 and Materials and Methods for details). This mutant showed no altered phenotype under any of a number of tested conditions, including growth with different nitrogen sources and growth with low levels of phosphate. In the Anabaena genome, three other PsiE-like protein-encoding ORFs are present, namely, all4445, alr1752, and all4381. It is possible that the presence in Anabaena of Alr0613 homologs compensates for the lack of Alr0613, explaining the phenotype of strain CSE39 (alr0613). On the other hand, an inactivated version of narM was constructed by insertion of gene cassette C.S3 (see Fig. 1 and Materials and Methods for details). One clone, denoted CSE38, in which no wild-type copies of narM could be detected, was selected for further analysis.
The ability of strain CSE38 (narM::C.S3) to grow in liquid medium using nitrate or nitrite as the nitrogen source was examined. Because an Anabaena strain unable to utilize nitrate or nitrite may produce heterocysts and grow fixing N2, the percentage of total cells that were heterocysts was also examined. Tested as a control, filaments of wild-type Anabaena and mutant CSE38 (narM) grown in the absence of a source of combined nitrogen contained 9.80% and 9.12% heterocysts, respectively. Both wild-type Anabaena and strain CSE38 (narM) grew in nitrite-containing media, producing a low percentage of heterocysts (0.97% in the wild type and 0.41% in the mutant). However, in nitrate-containing media, strain CSE38 (narM) was able to grow healthily but produced heterocysts (9.20%), which was in contrast to the findings for wild-type Anabaena, which produced only a few heterocysts (0.88%) under these conditions. These results suggest that strain CSE38 (narM) is impaired in nitrate assimilation but not in nitrite assimilation and, additionally, that it is able to grow diazotrophically.
The nitrate reductase and nitrite reductase activities were then determined. In contrast to wild-type Anabaena, strain CSE38 (narM) exhibited only basal levels of nitrate reductase activity under all incubation conditions tested (Table 3). In addition, this strain showed reduced levels of nitrite reductase activity, mainly in nitrate-containing medium (5% of wild-type activity) but also to some extent in nitrite-containing medium (62% of wild-type activity). Thus, the inability of strain CSE38 (narM) to use nitrate as a nitrogen source could result from the very low levels of nitrate and nitrite reductase activities in this mutant. The very low levels of nitrate reductase activity observed are consistent with the idea that alr0614 represents the narM gene of Anabaena.
TABLE 3.
Nitrate reductase and nitrite reductase activities of Anabaena sp. strain PCC 7120 (wild type) and mutant strains CSE38 (narM) and CSE40 (narB)a
| Strain | Enzymatic activity (mU · mg of protein−1) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nitrate reductase |
Nitrite reductase |
|||||||
| Nitrate | Nitrite | N2 | Ammonium | Nitrate | Nitrite | N2 | Ammonium | |
| PCC 7120 | 73.6 ± 9.0 | 46.1 ± 5.2 | 8.5 ± 1.3 | 7.2 ± 1.1 | 19.9 ± 1.6 | 6.9 ± 1.7 | 0.2 ± 0.4 | <0.1 |
| CSE38 | <0.1 | 1.5 ± 1.3 | <0.1 | 0.5 ± 0.8 | 1.0 ± 0.6 | 4.3 ± 0.4 | <0.1 | <0.1 |
| CSE40 | <0.1 | <0.1 | <0.1 | <0.1 | 0.2 ± 0.2 | 5.3 ± 0.8 | <0.1 | <0.1 |
Ammonium-grown cells were washed, resuspended in medium with the indicated source of nitrogen, and incubated for 4 h as described previously (10) for derepression experiments, and enzymatic activities were then determined. The values shown are the means and standard deviations from two to four independent experiments.
To discern whether the phenotype of strain CSE38 (narM) resulted from inactivation of narM itself or from the effect that this mutation has on nitrate reductase activity, we inactivated the nitrate reductase structural gene narB (ORF alr0612). A narB mutant, strain CSE40, was constructed by removing an internal 588-bp fragment to generate an in-frame deletion of the gene (the section encoding amino acid residues 222 to 417), thus minimizing possible polar effects on downstream genes (see Fig. 1 and Materials and Methods for details). As expected, strain CSE40 (narB) produced heterocysts in cultures lacking combined nitrogen (9.12%) and in nitrate-containing media (9.15%) but not in nitrite-containing media (0.21%), indicating its inability to use nitrate specifically as a nitrogen source. Nitrate and nitrite reductase activities were measured in strain CSE40 (narB). As expected, this strain presented negligible levels of nitrate reductase activity (Table 3). On the other hand, similar to strain CSE38 (narM), strain CSE40 (narB) showed decreased levels of nitrite reductase activity, mainly in nitrate-containing medium (1% of wild-type activity) but also somewhat in nitrite-containing medium (77% of wild-type activity). These results suggest that the phenotype of strain CSE38 (narM) resulted from the effect that the inactivation of narM has on the development of nitrate reductase activity. Additionally, these results indicate that a functional nitrate reductase is required for attaining high nitrite reductase levels when nitrate is present in the culture medium.
Nitrite reductase levels and nirA operon expression.
To study whether the reduced levels of nitrite reductase activity observed in mutant strains CSE38 (narM) and CSE40 (narB) corresponded to decreased amounts of the nitrite reductase protein, Western analysis was performed with polyclonal antibodies raised against Anabaena nitrite reductase (see Materials and Methods for details). In cell extracts from filaments of wild-type Anabaena incubated for 4 h under different nitrogen regimens, we detected a band that was not present in an nirA mutant (not shown) and whose levels were higher in nitrate- than in nitrite-containing media and the lowest in ammonium-containing media and in media without combined nitrogen (Fig. 4). This band was not present in cells from ammonium-containing established cultures of Anabaena (Fig. 4, lanes t0). In strains CSE38 (narM) and CSE40 (narB), decreased levels of the nitrite reductase protein were mainly observed in nitrate-containing media (Fig. 4). Therefore, the decreased levels of nitrite reductase activity observed in these mutants could correspond to low levels of nitrite reductase protein.
FIG 4.
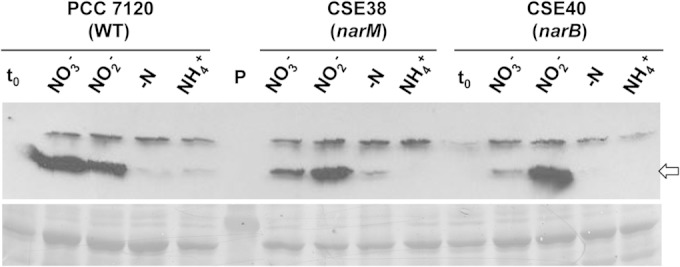
Detection of nitrite reductase protein in Anabaena strains. (Top) Western blot analysis with anti-NirA antibody and samples of strains PCC 7120, CSE38 (narM), and CSE40 (narB) grown in ammonium-containing medium (t0) or grown in ammonium-containing medium and incubated in medium with the indicated source of combined nitrogen: nitrate (NO3−), nitrite (NO2−), no combined nitrogen (−N), or ammonium (NH4+). Lane P, size markers. The arrow points to the nitrite reductase protein band. (Bottom) Portion of the Coomassie blue-stained membrane used in panel A showing the appropriate loading in all lanes.
To investigate whether the low levels of nitrite reductase detected in strains CSE38 (narM) and CSE40 (narB) resulted from an altered expression of the nirA operon, Northern analyses were carried out with probes for the nirA and narB genes. Hybridizations were performed with RNA isolated from filaments of wild-type Anabaena, CSE38 (narM), and CSE40 (narB) grown in ammonium and incubated in the presence of ammonium, nitrate, nitrite, or no combined nitrogen for 4 h. In wild-type Anabaena, a high level of expression of nirA took place in media without ammonium in the presence of nitrate or nitrite; however, it was higher in the presence of nitrate than in the presence of nitrite (Fig. 5). In the narM and narB mutants (strains CSE38 and CSE40, respectively), in contrast, a decreased level of expression of nirA compared to that in the wild type was observed, mainly in the presence of nitrate but also in the presence of nitrite. A similar expression profile was obtained with a probe corresponding to the 5′ end of narB (a fragment not deleted in CSE40; not shown). These data indicate that the decreased levels of nitrite reductase protein detected in strains CSE38 (narM) and CSE40 (narB) could result from a decreased level of expression of the nirA operon.
FIG 5.
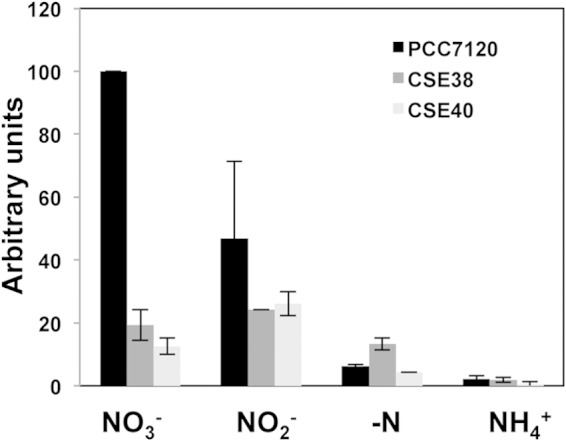
Expression of the nirA gene in strains PCC 7120 (wild type), CSE38 (narM), and CSE40 (narB). Hybridization assays were carried out using RNA isolated from cells grown with ammonium and incubated for 4 h in medium containing nitrate (NO3−), nitrite (NO2−), ammonium (NH4+), or no combined nitrogen (−N). The signals obtained were normalized with respect to those obtained with a probe of the rnpB gene. Data are in reference to the level of nirA expression for PCC 7120 in nitrate, which is considered 100%, and summarize the results of two to four experiments with independent cultures.
Since expression of the Anabaena nirA operon is dependent on two specific positive-action regulatory factors, NtcB and CnaT, expression of the genes encoding these regulators was also analyzed in strains CSE38 (narM) and CSE40 (narB). Expression profiles similar to those previously reported (22, 25) were found (not shown), indicating that the altered levels of nirA operon expression observed in strains CSE38 (narM) and CSE40 (narB) do not result from an abnormal expression of the regulatory gene ntcB or cnaT.
BACTH analysis of interactions between nitrate assimilation proteins.
For NarM of S. elongatus, a possible role in assembly of the different cofactors (iron-sulfur center and molybdopterin guanine dinucleotide) of nitrate reductase has been suggested (11). Our results also suggest a role for NarM in the production of a functional nitrate reductase in Anabaena. Additionally, in Anabaena, as in S. elongatus, NirB could act as a scaffolding protein for nitrite reductase (9, 10). To test whether these protein interactions are feasible, we used the bacterial two-hybrid (BACTH) system, which permits a visual screening for protein interactions on X-Gal-containing plates and a quantitative estimation of such interactions by determining β-galactosidase levels (40). Fusions of Anabaena NirB or NirA (nitrite reductase) to the C termini of T18 and T25 (the two complementary fragments of the catalytic domain of Bordetella pertussis adenylate cyclase), denoted T18-NirB, T18-NirA, T25-NirB, and T25-NirA, were prepared. Whereas the results of all control tests with one of these proteins and nonfused T25 or T18 were negative (β-galactosidase activity above background levels was not produced), strong interactions between NirB and itself and, especially, between nitrite reductase and NirB were observed (Fig. 6). The nitrite reductase-NirB interaction was observed independently of which of these proteins was fused to T18 or T25, and these results corroborate the previously proposed interaction between NirB and nitrite reductase (10).
FIG 6.
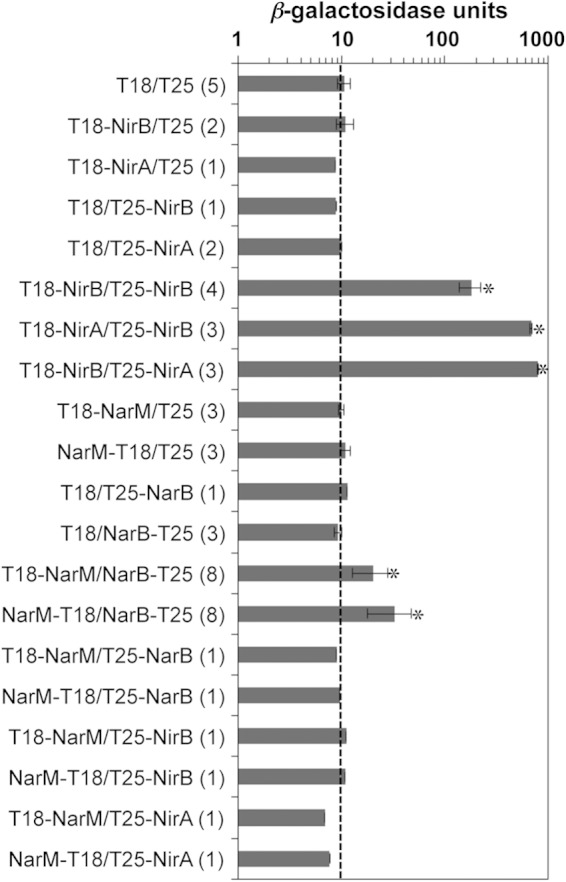
BACTH assays of interactions between Anabaena nitrate assimilation proteins. The interactions of the proteins fused to the adenylate cyclase T18 and T25 fragments cloned in E. coli were measured as the amount of β-galactosidase activity in liquid cultures as described in Materials and Methods and expressed as nanomoles of ONP per milligram of protein per minute. The protein fused to the N or the C terminus of T18 or T25 is indicated in each case (N terminus, protein-T18 or protein-T25; C terminus, T18-protein or T25-protein). The number of independent clones analyzed for each plasmid combination is shown in parentheses. An E. coli strain containing plasmids producing nonfused T18 and T25 was used as a general negative control and showed activity of 10.65 ± 1.45 nmol ONP mg protein−1 min−1 (mean ± SD). Controls with each T18 or T25 fusion protein and nonfused T25 or T18 were also run. All negative controls together rendered a value of 10.14 ± 1.33 nmol ONP mg protein−1 min−1 (mean ± SD). The vertical dotted line marks this background activity. Error bars reflect SDs (only when the number of assays indicated in parentheses was ≥2). *, the difference between the activity obtained with the indicated plasmid pair and the combined activity from all negative controls was significant at a P value of <0.01, as assessed by the Student t test.
To investigate possible interactions between NarB (nitrate reductase) and NarM, fusions of NarM to the N or C terminus of T18 (NarM-T18 and T18-NarM, respectively) and of NarB to the N or C terminus of T25 (NarB-T25 and T25-NarB, respectively) were prepared, and E. coli strains carrying the corresponding plasmid pairs and controls producing nonfused T25 or T18 were used in β-galactosidase assays. All pairs including a plasmid producing a nonfused T25 or T18 were negative, as expected (Fig. 6). In contrast, although the interactions were weaker than those between nitrite reductase and NirB or than NirB self-interactions, significant interactions between NarM and nitrate reductase could be observed with the T18-NarM/NarB-T25 and NarM-T18/NarB-T25 pairs but not when NarB was fused to the C terminus of T25 (Fig. 6). No interaction of NarM with NirB or NirA was detected, giving support to the specificity of the observed interactions of NarM with nitrate reductase. The low levels of β-galactosidase produced in the NarM/nitrate reductase tests might reflect transient interactions between NarM and nitrate reductase, in contrast to a more stable interaction that could take place between NirB and nitrite reductase.
DISCUSSION
Nitrate assimilation gene clusters including structural and regulatory genes are frequently found in cyanobacteria, and these gene clusters frequently contain an operon, the nirA operon, that consists of the structural genes for nitrite reductase, a nitrate transporter, and nitrate reductase. Here we have shown by Northern analysis that transcription of the nirA operon in Anabaena can extend to include ORF alr0613, which appears to be coexpressed with the nirA operon, at least when the Anabaena cells are incubated in the presence of nitrate. Indeed, RT-PCR analysis confirmed the coexpression of alr0613 and alr0612 (narB). Regarding alr0614 (narM), this gene exhibits an expression pattern similar to that shown by the nirA operon, but its coexpression with alr0612 (narB) and alr0613 could not be confirmed by RT-PCR. The correlated expression of narM and the nirA operon is discussed in the next paragraph. Nonetheless, a certain level of expression of alr0613 and narM appears to take place independently of the nirA operon, as observed in strain CSE36, in which transcription of the nirA operon is blocked at the level of nrtB (Fig. 2). Possible transcription start sites are located 29 bp upstream from the start codon of alr0613 and 48 bp upstream from the start codon of narM (41).
The correlated expression of narM and the nirA operon could hypothetically reflect the fact that the nirA operon includes narM, with processing of the nirA operon transcript producing an independently detectable narM transcript showing the same expression pattern as the nirA operon. Alternatively, transcription coming from alr0613 could promote expression from the narM promoter, thus leading to a coordinated expression of narM with the nirA operon. Cooperation in transcription has been proposed for the activation of two downstream promoters in a zinc-responsive operon of Anabaena (42) and might be based on antiroadblock mechanisms in transcription (43). Finally, the similar pattern of expression observed for narM and the nirA operon could involve the regulation of independent promoters by the same set of transcription factors. No binding site for NtcA or NtcB could, however, be identified in the sequences just upstream from alr0614 (narM).
Whereas ORF alr0613 does not appear to be necessary for nitrate assimilation under laboratory conditions in Anabaena, alr0614 represents the narM gene of Anabaena, which is necessary to develop nitrate reductase activity. Because a cyanobacterial narB gene alone provides E. coli with nitrate reductase activity (44), NarM does not appear to be a catalytic subunit of nitrate reductase. Instead, NarM appears to support the maturation of NarB (11). The coordinated expression of narM and the nirA operon would ensure the simultaneous production of NarM and the nitrate reductase enzyme, NarB, thus facilitating functional interactions between the two proteins, which may involve transient protein-protein interactions, as suggested by BACTH analysis.
The coordinated expression of narM and the nirA operon observed in Anabaena is in contrast to the situation in S. elongatus, in which narM is constitutively expressed at low levels (11). This may be related to the nitrate-dependent induction of the nirA operon, which is more stringent in Anabaena than in S. elongatus (see the introduction). In Anabaena, the nitrite reductase protein (NirA) and NirB are required to keep a low level of expression of the nirA operon in the absence of nitrate or nitrite (10). Here we have found that the two proteins interact strongly in BACTH analysis, confirming their joint role in nitrate assimilation, which could involve stable interactions between the two proteins under certain conditions. We have also observed that a functional nitrate reductase, which requires both the narB and narM genes to be produced, is needed to attain high levels of expression of the nirA operon in the presence of nitrate or nitrite. The requirement for narB and narM is, however, more stringent in the nitrate- than in the nitrite-dependent induction of the nirA operon (Fig. 5), which can also be clearly visualized in the production of the nitrite reductase protein (Fig. 4) and activity (Table 3). Hence, nitrite resulting from nitrate reduction (catalyzed by nitrate reductase) is a likely inducer of the nirA operon. This is consistent with the proposed role of nitrite in induction of the nirA operon involving NtcB in S. elongatus cells treated with l-methionine-d,l-sulfoximine (20). Exogenously added nitrite is, however, a poorer inducer of the nirA operon than nitrite produced inside the cells by a functional nitrate reductase (Table 3 and Fig. 5). This could result from a toxic effect of nitrite, but the possibility that an additional effector(s) related to the nitrate reductase reaction is involved in induction can also be raised. On the other hand, the basis for the requirement of narB and narM for attaining maximal levels of nitrite-dependent expression of the nirA operon (Fig. 5) is unknown.
In summary, we have shown that Anabaena bears a narM gene that is transcribed in a coordinated manner with the nirA operon and that narM, along with the nitrate reductase structural gene narB, is needed to produce nitrate reductase activity. Both narB and narM are also needed for nitrate-dependent induction of the nirA operon, suggesting that nitrite is an inducer. As previously shown (10), this nitrate/nitrite effect on the expression of the nitrate assimilation operon appears to involve a negative transcriptional effect of nitrite reductase (which also requires NirB) when there is no nitrite in the cytoplasm. The putative interactions NirA/NirB and NarB/NarM were tested here with the BACTH system, which has shown a strong interaction between nitrite reductase (NirA) and NirB and a weak interaction, perhaps indicative of transient interactions, between nitrate reductase (NarB) and NarM.
ACKNOWLEDGMENTS
We thank Antonia Herrero and Ignacio Luque (CSIC, Seville, Spain) for useful discussions and Félix Ramos-León, Ana Valladares, and Rocío Rodríguez (CSIC, Seville, Spain) for help with β-galactosidase assays, induction experiments, and purification of nitrite reductase, respectively.
This work was supported by grants BFU2008-03811 and BFU2011-22762 from the Plan Nacional de Investigación, Spain, cofinanced by the European Regional Development Fund.
REFERENCES
- 1.Moreno-Vivián C, Flores E. 2007. Nitrate assimilation in bacteria, p 263–282. In Bothe H, Ferguson SJ, Newton WE (ed), Biology of the nitrogen cycle. Elsevier B.V., Amsterdam, Netherlands. [Google Scholar]
- 2.Muro-Pastor MI, Reyes JC, Florencio FJ. 2005. Ammonium assimilation in cyanobacteria. Photosynth Res 83:135–150. doi: 10.1007/s11120-004-2082-7. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki A, Knaff DB. 2005. Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth Res 83:191–217. doi: 10.1007/s11120-004-3478-0. [DOI] [PubMed] [Google Scholar]
- 4.Flores E, Frías JE, Rubio LM, Herrero A. 2005. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth Res 83:117–133. doi: 10.1007/s11120-004-5830-9. [DOI] [PubMed] [Google Scholar]
- 5.Ohashi Y, Shi W, Takatani N, Aichi M, Maeda S, Watanabe S, Yoshikawa H, Omata T. 2011. Regulation of nitrate assimilation in cyanobacteria. J Exp Bot 62:1411–1424. doi: 10.1093/jxb/erq427. [DOI] [PubMed] [Google Scholar]
- 6.Flores E, Herrero A. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol 8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 7.Omata T. 1995. Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC7942. Plant Cell Physiol 36:207–213. [DOI] [PubMed] [Google Scholar]
- 8.Frías JE, Flores E, Herrero A. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 179:477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki I, Horie N, Sugiyama T, Omata T. 1995. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J Bacteriol 177:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frías JE, Flores E. 2010. Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 192:2769–2778. doi: 10.1128/JB.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda S-I, Omata T. 2004. A novel gene (narM) required for expression of nitrate reductase activity in the cyanobacterium Synechococcus elongatus strain PCC 7942. J Bacteriol 186:2107–2114. doi: 10.1128/JB.186.7.2107-2114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero A, Flores E, Guerrero MG. 1981. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J Bacteriol 145:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero A, Muro-Pastor AM, Flores E. 2001. Nitrogen control in cyanobacteria. J Bacteriol 183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanigawa R, Shirokane M, Maeda Si S, Omata T, Tanaka K, Takahashi H. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc Natl Acad Sci U S A 99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vázquez-Bermúdez MF, Herrero A, Flores E. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett 512:71–74. doi: 10.1016/S0014-5793(02)02219-6. [DOI] [PubMed] [Google Scholar]
- 16.Valladares A, Flores E, Herrero A. 2008. Transcription activation by NtcA and 2-oxoglutarate of three genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 190:6126–6133. doi: 10.1128/JB.00787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao MX, Jiang YL, He YX, Chen YF, Teng YB, Chen Y, Zhang CC, Zhou CZ. 2010. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc Natl Acad Sci U S A 107:12487–12492. doi: 10.1073/pnas.1001556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinosa J, Forchhammer K, Burillo S, Contreras A. 2006. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol 61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 19.Llácer JL, Espinosa J, Castells MA, Contreras A, Forchhammer K, Rubio V. 2010. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc Natl Acad Sci U S A 107:15397–15402. doi: 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aichi M, Omata T. 1997. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol 179:4671–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aichi M, Takatani N, Omata T. 2001. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183:5840–5847. doi: 10.1128/JB.183.20.5840-5847.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frías JE, Flores E, Herrero A. 2000. Activation of the Anabaena nir operon promoter requires both NtcA (CAP family) and NtcB (LysR family) transcription factors. Mol Microbiol 38:613–625. doi: 10.1046/j.1365-2958.2000.02156.x. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto T, Inoue-Sakamoto K, Persson S, Bryant DA. 2008. Transcription factor NtcB specifically controls the nitrate assimilation genes in the marine cyanobacterium Synechococcus sp. strain PCC 7002. Phycol Res 56:223–237. doi: 10.1111/j.1440-1835.2008.00504.x. [DOI] [Google Scholar]
- 24.Kikuchi H, Aichi M, Suzuki I, Omata T. 1996. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol 178:5822–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frías JE, Herrero A, Flores E. 2003. Open reading frame all0601 from Anabaena sp. strain PCC 7120 represents a novel gene, cnaT, required for expression of the nitrate assimilation nir operon. J Bacteriol 185:5037–5044. doi: 10.1128/JB.185.17.5037-5044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 27.Ausubel FM, Brent R, Kingston RE, Moore D, Seidman JG, Smith JA, Struhl K. 2009. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY. [Google Scholar]
- 28.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 31.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol 179:1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muro-Pastor AM, Valladares A, Flores E, Herrero A. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol Microbiol 44:1377–1385. doi: 10.1046/j.1365-2958.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- 33.Church GM, Gilbert W. 1984. Genomic sequencing. Proc Natl Acad Sci U S A 81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markwell MAK, Hass SM, Bieber LL, Tolbert NE. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem 87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 35.Luque I, Andújar A, Jia L, Zabulon G, Tandeau de Marsac N, Flores E, Houmard J. 2006. Regulated expression of glutamyl-tRNA synthetase is directed by a mobile genetic element in the cyanobacterium Tolypothrix sp. PCC 7601. Mol Microbiol 60:1276–1288. doi: 10.1111/j.1365-2958.2006.05170.x. [DOI] [PubMed] [Google Scholar]
- 36.Herrero A, Flores E, Guerrero MG. 1985. Regulation of nitrate reductase cellular levels in the cyanobacteria Anabaena variabilis and Synechocystis sp. FEMS Microbiol Lett 26:21–25. doi: 10.1111/j.1574-6968.1985.tb01559.x. [DOI] [Google Scholar]
- 37.Herrero A, Guerrero MG. 1986. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J Gen Microbiol 132:2463–2468. [Google Scholar]
- 38.Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 39.Elhai J, Wolk CP. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 40.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5256. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. 2011. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A 108:20130–20135. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napolitano M, Rubio MA, Camargo S, Luque I. 2013. Regulation of internal promoters in a zinc-responsive operon is influenced by transcription from upstream promoters. J Bacteriol 195:1285–1293. doi: 10.1128/JB.01488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. 2003. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J 22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubio LM, Herrero A, Flores E. 1996. A cyanobacterial narB gene encodes a ferredoxin-dependent nitrate reductase. Plant Mol Biol 30:845–850. doi: 10.1007/BF00019017. [DOI] [PubMed] [Google Scholar]
- 45.Vioque A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res 25:3471–3477. doi: 10.1093/nar/25.17.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8:205–213. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 47.Wolk CP, Cai Y, Cardemil L, Flores E, Hohn B, Murry M, Schmetterer G, Schrautemeier B, Wilson R. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J Bacteriol 170:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muro-Pastor AM, Valladares A, Flores E, Herrero A. 1999. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol 181:6664–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol 9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]