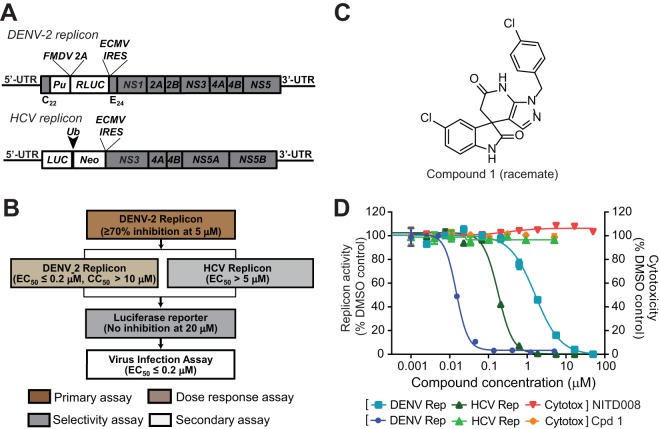
FIG 1.
(A) Schematic diagram of the Renilla luciferase (RLUC)-expressing DENV-2 replicon (New Guinea C strain) and the firefly luciferase (LUC)-expressing HCV replicon. The diagram is not drawn to scale. C22, N-terminal 22 amino acids of the DENV-2 capsid protein; E24, C-terminal 24 amino acids of the E protein; Pu, puromycin N-acetyltransferase gene; FMDV 2A, foot-and-mouth disease virus 2A protein; IRES, internal ribosomal entry site from encephalomyocarditis virus; Ub, ubiquitin; Neo, neomycin resistance gene; UTR, untranslated region. (B) Dual-replicon screening flow chart. Compound libraries were screened at 5 μM by using Renilla luciferase DENV-2 replicon cells. Compounds with >70% inhibition of Renilla luciferase activity in the primary screening were subjected to a dose-response test in Renilla luciferase DENV-2 replicon cells to derive EC50 and CC50 values. The compounds were also tested against the HCV firefly luciferase replicon. Compounds that selectively inhibited the DENV-2 replicon were further examined in a Renilla luciferase reporter cell line to rule out Renilla luciferase inhibitors. The inhibitors were further validated in a viral titer reduction assay using authentic DENV. (C) Chemical structure of compound 1 (racemate). (D) Antiviral activities of compound 1 (Cpd 1). NITD-008, a nucleoside inhibitor of DENV and HCV (12), was included as a positive control. DENV-2 Pu-Renilla luciferase replicon BHK-21 cells or HCV Neo-firefly luciferase replicon Huh-7 cells were cultured in medium containing 3-fold serial dilutions of compound 1 for 2 days at 37°C. ViviRen (Promega) and Britelite (PerkinElmer) were added to the DENV-2 and HCV replicons, respectively. The luciferase signal was detected with a Clarity 4.0 plate reader, followed by cell viability detection by the addition of CellTiter-Glo (Promega). The luciferase assay and the cell viability assay were performed according to the manufacturers' protocols. Antiviral activity and cell viability are presented as the left y axis and right y axis, respectively.