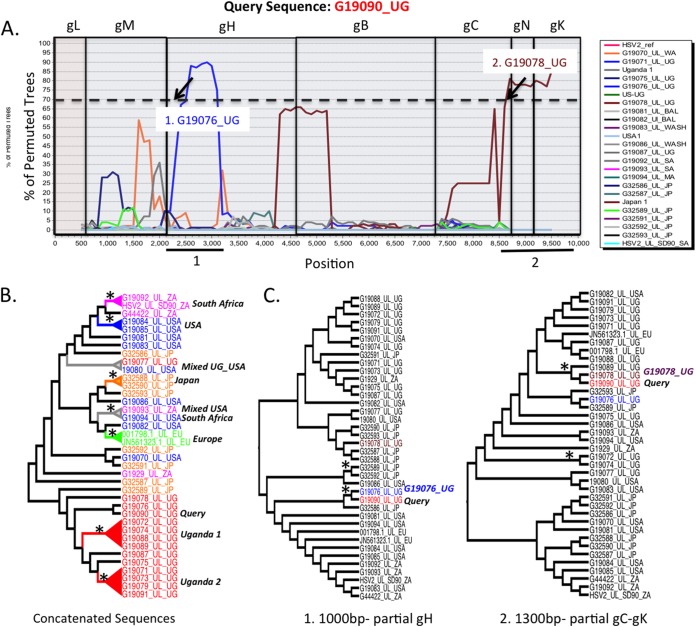
FIG 3.
Boot-scanning and phylogenetic analyses for evidence of recombination within concatenated HSV-2 UL glycoprotein sequences. (A) HSV-2 Uganda strain G19090_UG was used as the query sequence against other HSV-2 UL concatenated glycoprotein sequences using a boot-scanning approach. The x axis reflects the position in the aligned set of sequences, and the y axis shows the percentage of permuted trees in which an individual HSV-1 strain clustered with the query. A recombination cutoff value of 70% is indicated by the dashed line. Regions of the scan associated with each glycoprotein segment are boxed, with the glycoprotein name above the plot. Two putative recombination events occurred and are numbered. The arrows indicate positions where sequence concatenation may be linked to the recombination signal. Directly below the boot-scanning plot, numbered black lines indicate regions of the alignment used to substantiate recombination signal in the phylogenies shown in panel C. (B) Concatenated glycoprotein phylogeny (far left) with taxa colored by region (green, Europe; blue, United States; red, Africa; orange, Asia). The tree was used as a guide to group some sequences in the boot-scanning analysis plot shown in panel A. Boot-scanning groups, indicated by colored cones subtending the sequence names, were defined by bootstrap significance of >90% and included two African groups (1 and 2), one European group, one Asian group, one U.S. group, one South African group, and two mixed groups containing sequences from (i) Uganda and the United States and (ii) South Africa and the United States. (C) Two midpoint-rooted phylogenies were generated and are shown in cladogram format. These phylogenies were generated with segments of the alignment as noted in panel A. The lengths of the segments used to create the phylogeny are shown at the bottom. The strain containing the most closely related recombinant segment is indicated next to the phylogeny with the query sequence (G19090_UG). The asterisks indicate bootstrap values of 70% or higher.