FIG 1.
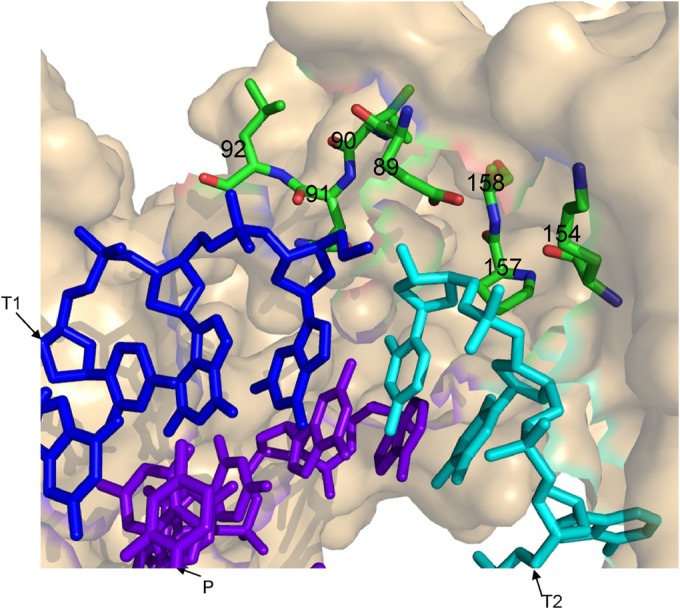
3D model for HIV-1 RT and clamp substrates. Using PyMol (PyMOL Molecular Graphics System, v1.4.1; Schrödinger, LLC), and based on 1ROA HIV-1 RT crystal structure (28), residues closer than 7 Å to the clamp site have been marked (i.e., residues 89 to 92, 154, 157, and 158 of the p66 RT subunit are highlighted and numbered). Blue, T1 (the first template); cyan, T2 (the second template); purple, P (the primer). Energy minimization was performed with the GROMACS 4.0.7 software suite, using the steepest descent algorithm (29).
