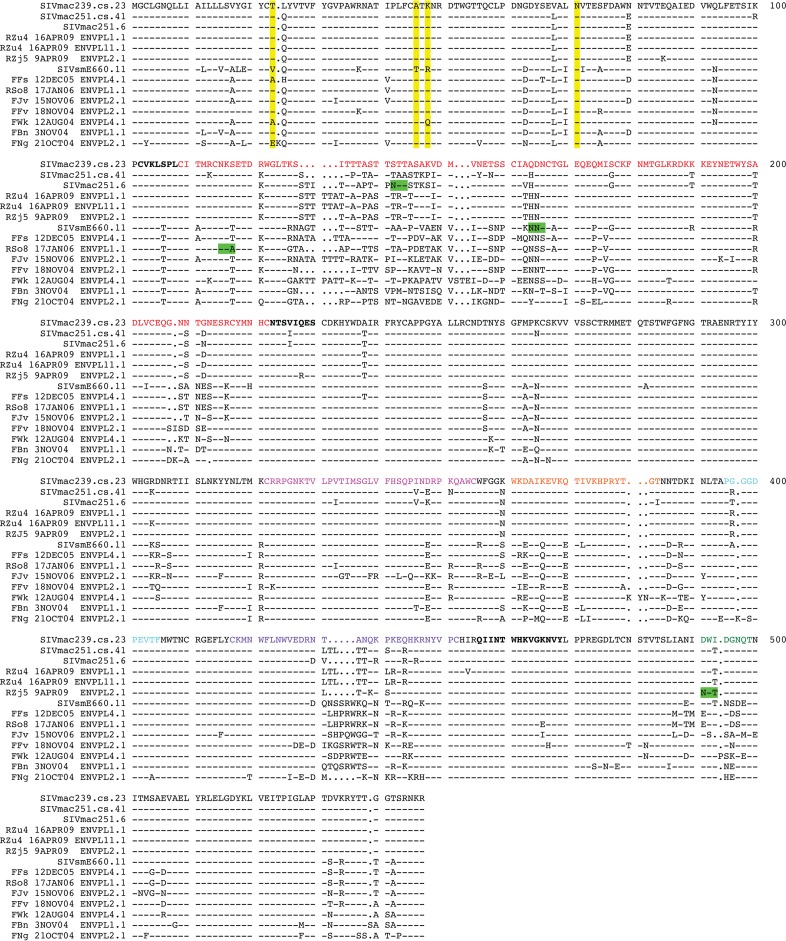
FIG 3.
Amino acid sequence alignment of the SIVsm Env panel. An amino acid alignment of the Env gp120-coding sequence was generated using the SeqPublish tool, available from the Los Alamos National Laboratory HIV Database (http://www.hiv.lanl.gov/content/sequence/SeqPublish/seqpublish.html). The sequence of SIVmac239.cs.23 is shown as the reference sequence, and dashes indicate conserved residues, while dots indicate deleted residues. Yellow highlighting, the positions of the four amino acid residues (positions 23, 45, 47, and 70) described in reference 18, with A45 and K47 being described to be major determinants of SIVsmE660 neutralization resistance in that reference; green highlighting, atypical PNLG sites that are associated with the tier 1 Envs. Functional gp120 domains are indicated by colored text as follows: red, V1V2; magenta,V3; orange, α2 helix; purple, V4; green,V5; bold text, bridging sheet residues, as described previously (72); cyan, the CD4 binding loop.