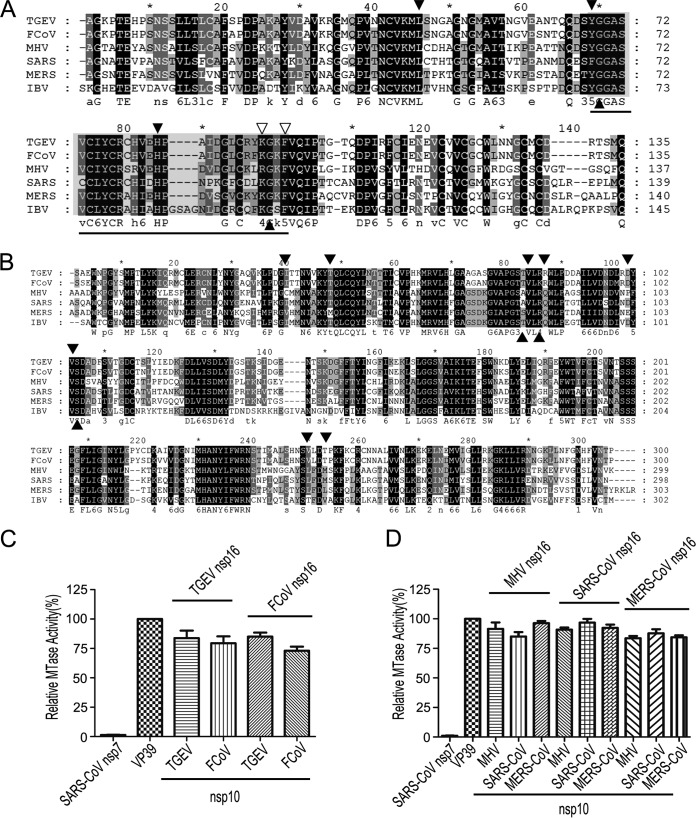
FIG 3.
Conservation of nsp10 and nsp16 in coronaviruses. (A and B) Primary sequence alignments of nsp10 (A) and nsp16 (B) of TGEV, FCoV, MHV, SARS-CoV, MERS-CoV, and IBV. The well-conserved and relatively conserved residues are indicated by solid black boxes and solid gray boxes, respectively. The conserved residues involved in the interactions of nsp10/nsp16 complexes are indicated by solid black and open triangles. The region corresponding to the peptide inhibitor P29 is marked by a line. The alignment was generated with the program Clustal, and the figure was prepared using Genedoc. (C and D) MTase assays of alphacoronaviral nsp16 (C) and betacoronaviral nsp16 (D) stimulated by the noncognate nsp10 from a coronavirus of the same genus. SARS-CoV nsp7 and vaccinia virus 2′-O-MTase (VP39) were used as a negative and positive control, respectively (n = 3; mean values ± SD).