FIG 2.
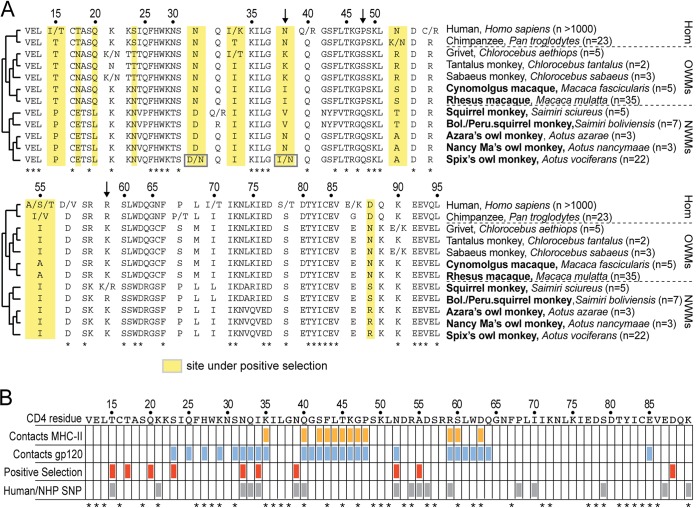
There are many nonsynonymous SNPs in the portion of CD4 encoding the D1 domain. (A) An amino acid alignment of the D1 domain from species included in the population study. Nonsynonymous SNPs identified are indicated with slashes, where the two alternately encoded amino acids are given on either side. Dotted lines separate the major clades of simian primates (Hom, hominoids; OWMs, Old World monkeys; NWMs, New World monkeys). The number of individuals analyzed from each species is shown in parentheses adjacent to the species name. Amino acid positions highlighted in yellow were previously identified to be evolving under positive selection (3). Numbering along the top is relative to the mature CD4 protein, after cleavage of the 25-amino-acid N-terminal signal peptide. Species shown in bold indicate populations that were sequenced in this study. Arrows indicate three sites that have been shown previously to affect HIV-1 entry (14, 17). (B) A table showing amino acids in the CD4 D1 domain that contact gp120 (blue) or MHC-II (orange), are under positive selection (red) (3), or are polymorphic for nonsynonymous mutations in human or nonhuman primate (NHP) populations included in the present study (gray). Asterisks along the bottom denote residue positions that are completely conserved in the 31 primate species shown in Fig. 1C. The table format is modified from reference 4.