Abstract
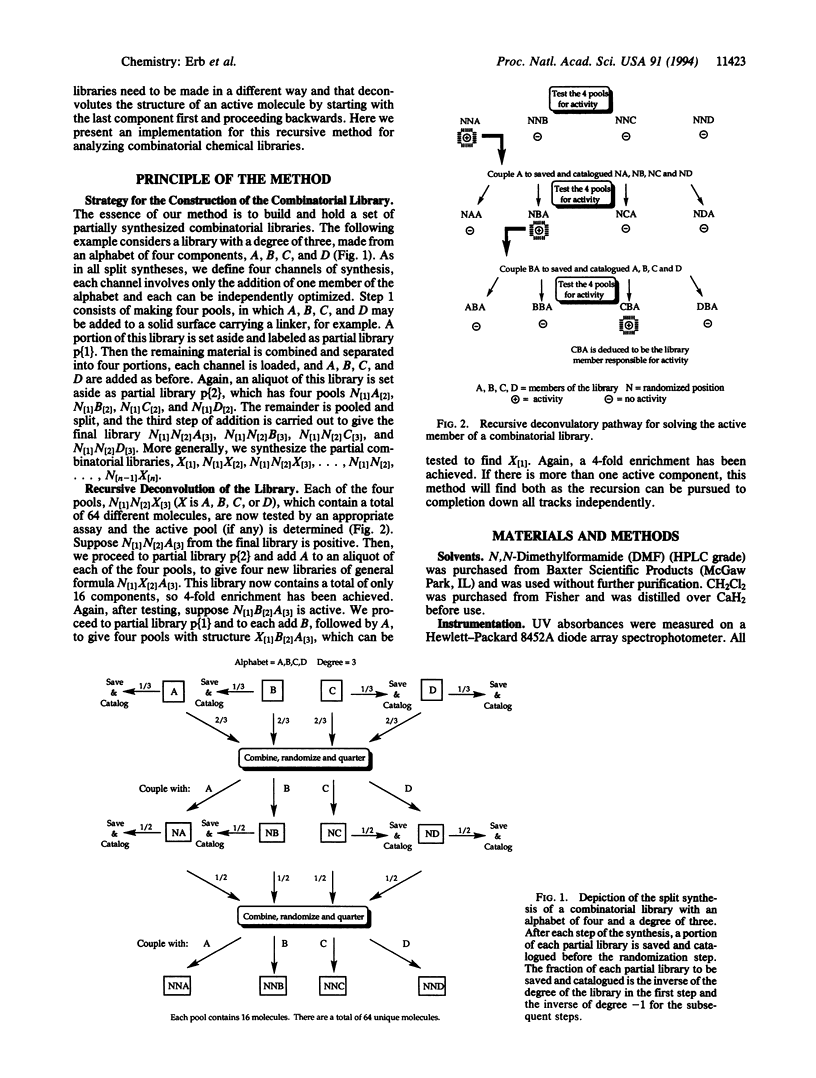
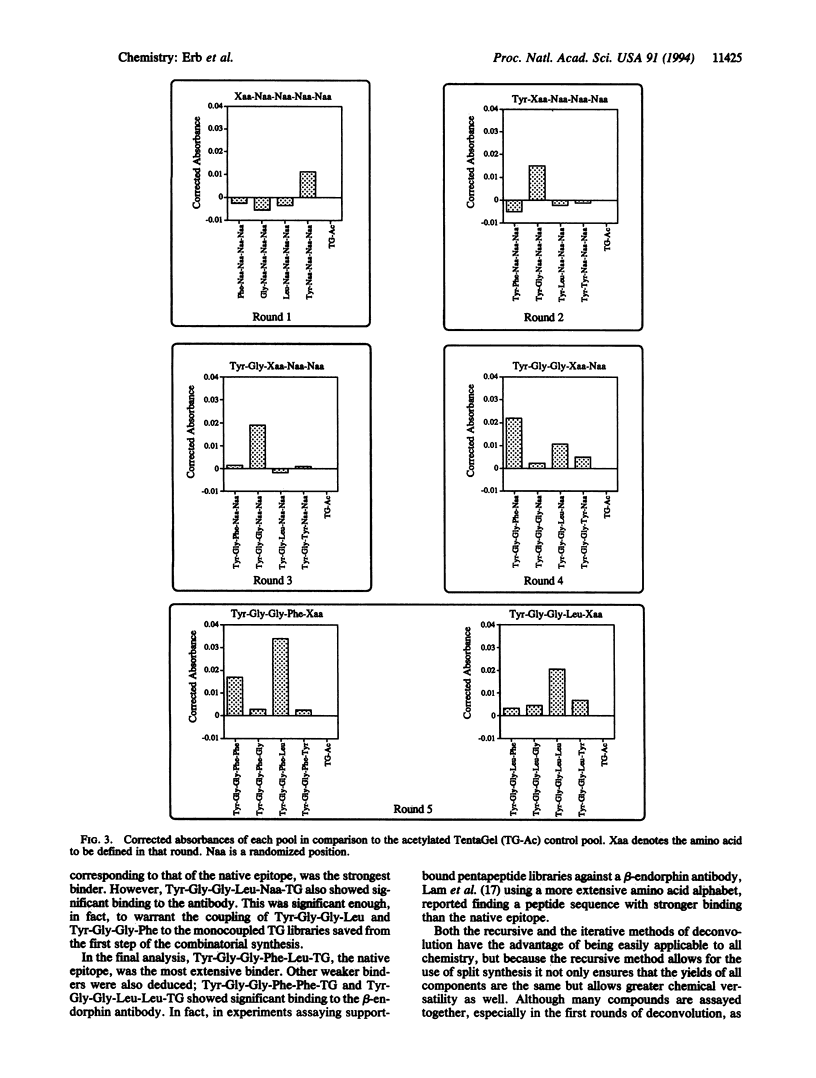
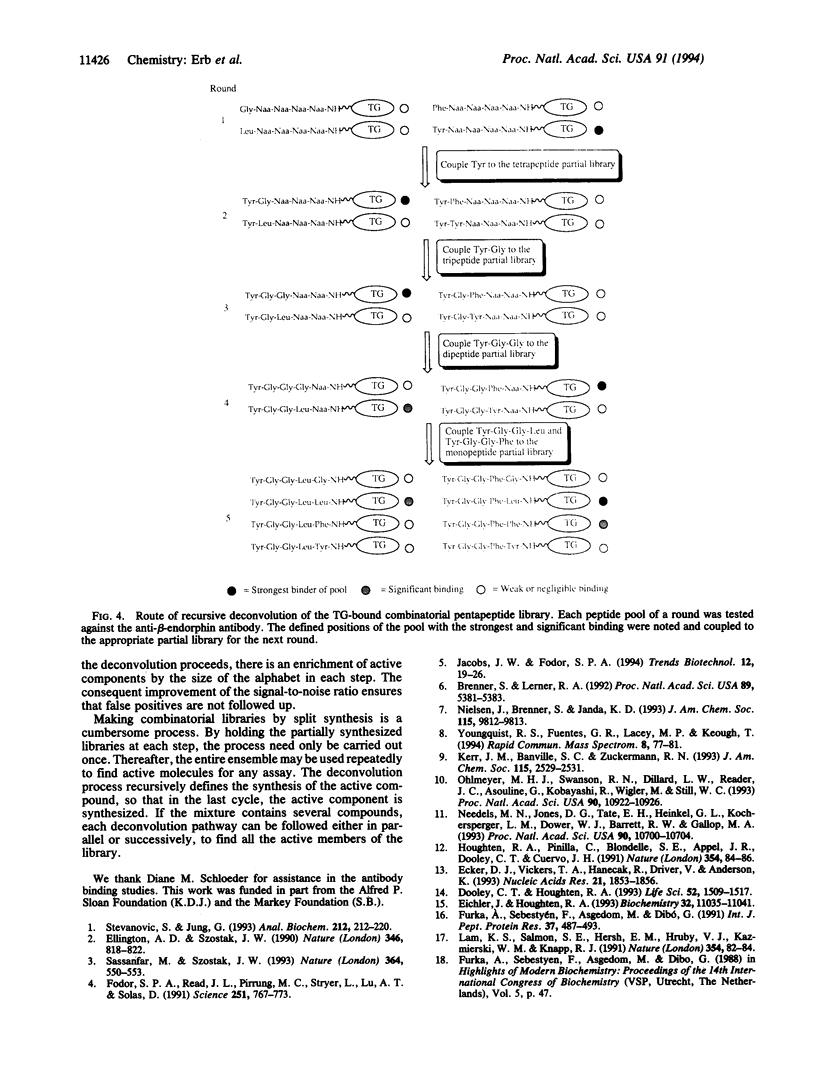
A recursive strategy that solves for the active members of a chemical library is presented. A pentapeptide library with an alphabet of Gly, Leu, Phe, and Tyr (1024 members) was constructed on a solid support by the method of split synthesis. One member of this library (NH2-Tyr-Gly-Gly-Phe-Leu) is a native binder to a beta-endorphin antibody. A variation of the split synthesis approach is used to build the combinatorial library. In four vials, a member of the library's alphabet is coupled to a solid support. After each coupling, a portion of the resin from each of the four reaction vials was set aside and catalogued. The solid support from each vial is then combined, mixed, and redivided. The steps of (i) coupling, (ii) saving and cataloging, and (iii) randomizing were repeated until a pentapeptide library was obtained. The four pentapeptide libraries where the N-terminal amino acid is defined were screened against the beta-endorphin antibody and quantitated via an ELISA. The amino acid of the four pools that demonstrated the most binding was then coupled to the four tetrapeptide partial libraries that had been set aside and catalogued during the split synthesis. This recursive deconvolution was repeated until the best binders were deduced. Besides the anticipated native binder, two other members of the library displayed significant binding. This recursive method of deconvolution does not use a molecular tag, requires only one split synthesis, and can be applied to the deconvolution of nonlinear small-molecule combinatorial libraries and linear oligomeric combinatorial libraries, since it is based only on the procedure of the synthesis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner S., Lerner R. A. Encoded combinatorial chemistry. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley C. T., Houghten R. A. The use of positional scanning synthetic peptide combinatorial libraries for the rapid determination of opioid receptor ligands. Life Sci. 1993;52(18):1509–1517. doi: 10.1016/0024-3205(93)90113-h. [DOI] [PubMed] [Google Scholar]
- Ecker D. J., Vickers T. A., Hanecak R., Driver V., Anderson K. Rational screening of oligonucleotide combinatorial libraries for drug discovery. Nucleic Acids Res. 1993 Apr 25;21(8):1853–1856. doi: 10.1093/nar/21.8.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J., Houghten R. A. Identification of substrate-analog trypsin inhibitors through the screening of synthetic peptide combinatorial libraries. Biochemistry. 1993 Oct 19;32(41):11035–11041. doi: 10.1021/bi00092a013. [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991 Feb 15;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Furka A., Sebestyén F., Asgedom M., Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991 Jun;37(6):487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Houghten R. A., Pinilla C., Blondelle S. E., Appel J. R., Dooley C. T., Cuervo J. H. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991 Nov 7;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Jacobs J. W., Fodor S. P. Combinatorial chemistry--applications of light-directed chemical synthesis. Trends Biotechnol. 1994 Jan;12(1):19–26. doi: 10.1016/0167-7799(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Lam K. S., Salmon S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., Knapp R. J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991 Nov 7;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Needels M. C., Jones D. G., Tate E. H., Heinkel G. L., Kochersperger L. M., Dower W. J., Barrett R. W., Gallop M. A. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer M. H., Swanson R. N., Dillard L. W., Reader J. C., Asouline G., Kobayashi R., Wigler M., Still W. C. Complex synthetic chemical libraries indexed with molecular tags. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Szostak J. W. An RNA motif that binds ATP. Nature. 1993 Aug 5;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Stevanović S., Jung G. Multiple sequence analysis: pool sequencing of synthetic and natural peptide libraries. Anal Biochem. 1993 Jul;212(1):212–220. doi: 10.1006/abio.1993.1314. [DOI] [PubMed] [Google Scholar]
- Youngquist R. S., Fuentes G. R., Lacey M. P., Keough T. Matrix-assisted laser desorption ionization for rapid determination of the sequences of biologically active peptides isolated from support-bound combinatorial peptide libraries. Rapid Commun Mass Spectrom. 1994 Jan;8(1):77–81. doi: 10.1002/rcm.1290080115. [DOI] [PubMed] [Google Scholar]



