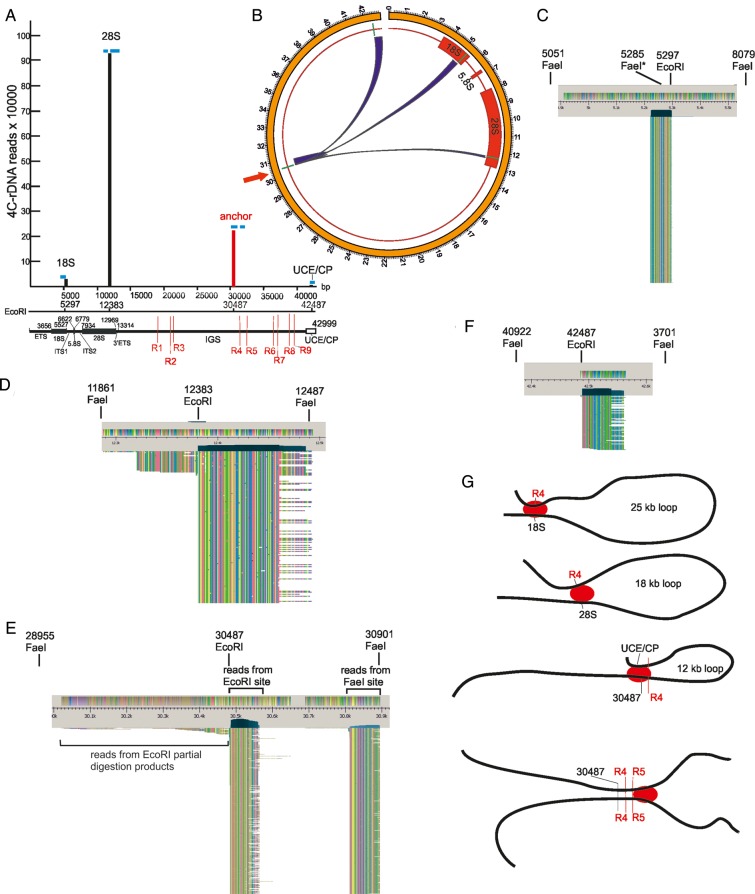
Figure 7.
Contacts inside and between rDNA units. (A) Vertical bars present the number of mapped 4C-rDNA reads mapped inside rDNA units. Blue bars indicate the regions where reads were mapped in respect to four EcoRI sites inside the rDNA unit. Thin red lines inside IGS show the positions of R1–R9 possessing hot spots of DSBs. The reads at coordinate 5297 were detected only to the left of the EcoRI site suggesting that the contacts involved only the fragment possessing ETS and the 5′ part of 18S gene, but not the region between coordinates 5297 and 12383. We detected only a small number of reads (∼1%) to the left of coordinate 12383, while the rest of them—∼1 million reads—were mapped exclusively to the right of the coordinate, indicating that this particular region at the 3′ end of 28S rDNA gene is very often located in close proximity to the anchor site (shown with a red vertical bar at coordinate 30487). We detected the reads on both sides around the anchor. The reads to the left of coordinate 30487 may result from both the partial digestion products during EcoRI treatment, and from the contacts with the neighboring 18-kb region repairing the site during ligation (see Figure 5A). The reads to the right of coordinate 30487 could appear only from the contacts with the same 12-kb fragment from another rDNA unit. These reads are mapped in two regions coming either from EcoRI and FaeI sites during inverse PCR. We did not observe any reads that potentially could appear after circularization of 12-kb EcoRI fragments possessing the 4C-anchor (to the left of coordinate 42487). The location of a small number of reads only to the right of EcoRI site at coordinate 42487 suggests that a small portion of rDNA units are observed in close proximity to the corresponding region to the anchor site (coordinate 30487). (B) Circos presentation of contacts inside the rDNA unit itself. EcoRI sites are shown by short, green lines. The contacts of the anchor site (shown by the red arrow) were detected inside the 18S and 28S genes, downstream from the anchor site, and in the region of the UCE/CP. (C) Overview of reads at the coordinate 5297. The sizes of EcoRI-FaeI fragments involved in the 4C procedure were 234 bp and 2.8 kb. The asterisk at one FaeI site indicates the diverged sequence. (D) Overview of reads at the coordinate 12383. The sizes of EcoRI-FaeI fragments involved in the 4C procedure were 522 bp and 104 bp. 99% of reads were detected to the right of the EcoRI site. (E) Overview of reads at the anchor EcoRI site (coordinate 30487). A small number of reads located to the left of the EcoRI site correspond to partial digestion products during EcoRI treatment (see Figure 5A). Reads coming from EcoRI and FaeI sites during inverse PCR are seen in two columns (only the top 596 rows are shown). They could originate only from the contacts between the same regions at the anchor EcoRI site from different rDNA units. (F) Overview of reads at the coordinate 42487. The sizes of EcoRI-FaeI fragments involved in the 4C procedure were 1.5 kb and 4.2 kb. Although the size of the latter fragment is rather large, the reads at this coordinate were detected only to the right of the EcoRI site, which strongly indicates that there was practically no circularization of 12-kb fragments (located between coordinates 30487 and 42487) during the 4C procedure. (G) Possible formation of loops inside the rDNA units and between them. Red ovals indicate the cross-linking proteins.