Fig. 1.
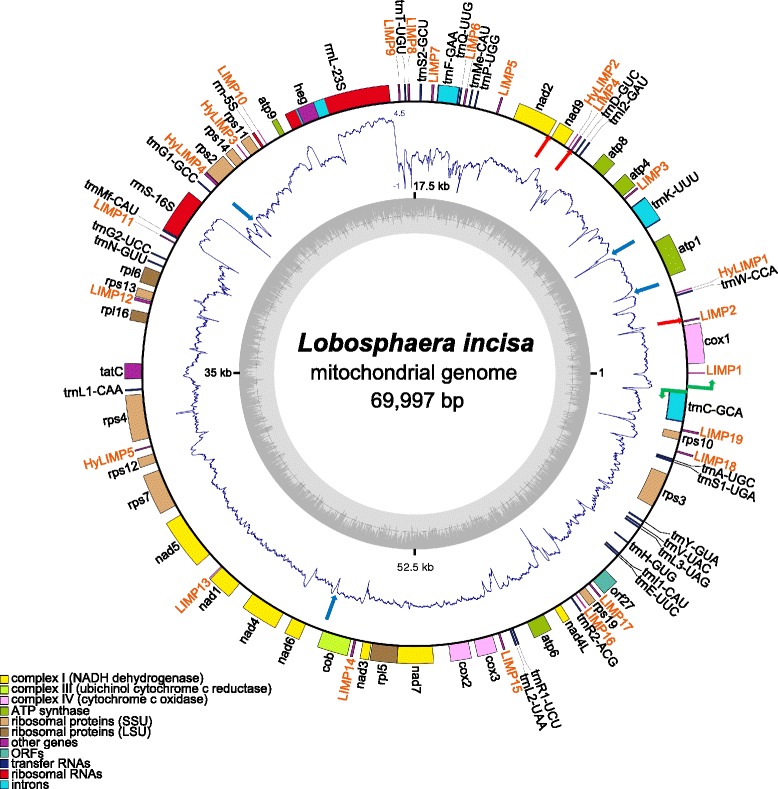
Schematic representation of the L. incisa mitochondrial genome. Genes depicted on the outside of the outer circle are transcribed in counterclockwise orientation, on the inside in clockwise orientation. Putative promoters are shown as green bent arrows. Genes are color-coded according to functional classes, as listed in the bottom left corner. The LIMP and HyLIMP copies are shown in orange. The inner ring displays a graph of the G + C content (the circle marks the 50 % threshold), the middle one the RNA-Seq coverage (log10 scale, with 0 values brought to −1). Red arrows indicate the locations of the three short regions that are deleted in a fraction of the molecules, and blue arrows indicate the locations of the four palindromes (not LIMPs) of high predicted stability that are associated with a drop or loss of RNA-Seq coverage. The figure was generated using OrganellarGenomeDRAW [74]. The coverage plot was drawn using the “polar.plot” function from the plotrix package in R
