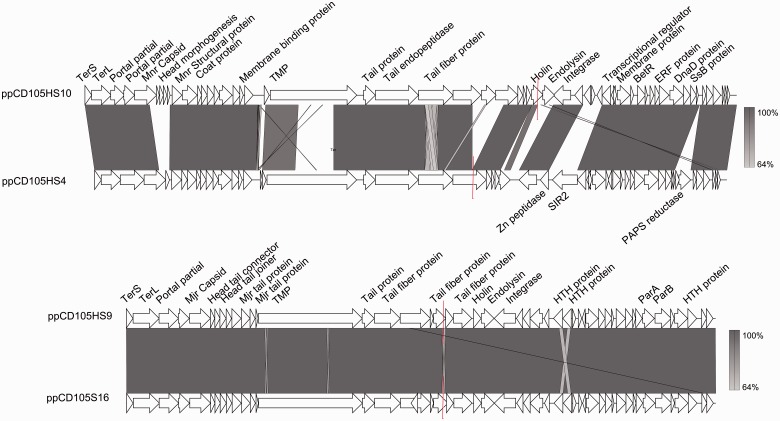
Fig. 3.—
Two novel prophage-like regions containing siphovirus-like CDSs. Linear genome maps of the predicted prophage regions. Top: CD105H9S10 (ppCD105HS10) and CD105HS4 (ppCD105HS4), below: CD105HS16 (ppCD105HS16) and CD105HS9 (ppCD105HS9). Isolates CD105HS10 and CD105HS4 are different ribotypes, R005 and R014, respectively, but CD105HS16 and CD105HS are both R010. The prophage regions have CDSs encoding phage proteins involved in all stages of temperate phage infecting, and are arranged in a similar architecture to the other C. difficile phages with terminase, structural, lysis and attachment, lysogeny control, and DNA replication. Color shading indicates sequence similarity through BLASTn, and red lines indicate a break in the region by contig gap.