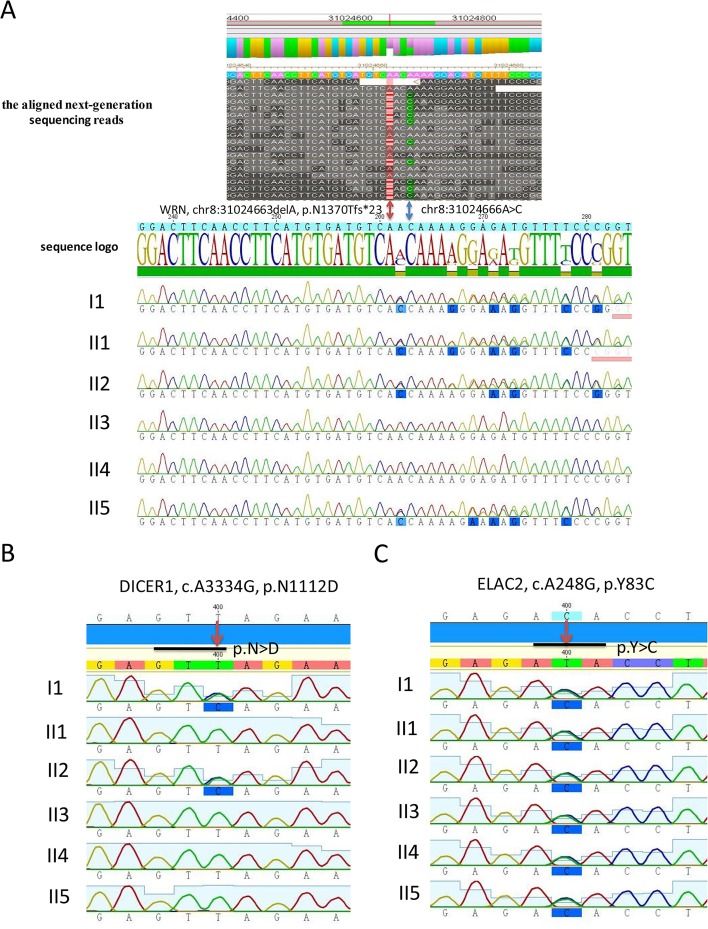
Fig 2. Sanger sequencing of germline mutations identified in WRN, DICER1, and ELAC2.
(A) Sanger sequencing validation of WRN frameshift mutation in each individual. The aligned NGS data of WRN mutation from the mother (I:1). The WRN frameshift mutation presented a 1-bp deletion in chr8:31024663. The bases after A are shown in red, and the A→C point mutation in chr8:31024666 is shown in blue. The WRN frameshift mutation c.4108DelA (p.N1370Tfs*23) was validated by Sanger sequencing in the mother (I:1), the proband (II:1), the second daughter (II:2), and the youngest daughter (II:5); however, it was absent in the other two daughters (II:3 and II:4). (B) The DICER1 missense mutation c.A3334G (p.N1112D) was validated by Sanger sequencing in the mother (I:1) and the second daughter (II:2), but it was absent in the other members of this family. (C) The ELAC2 mutation c.A248G (p.Y83C) was validated by Sanger sequencing in all the members of this family.