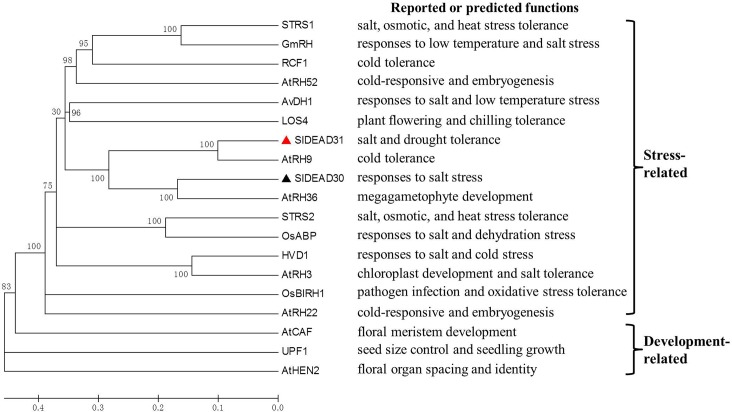
Fig 2. Phylogenetic tree and reported or predicted functions of SlDEAD30, SlDEAD31, and other known plant DEAD-box proteins.
The phylogenetic tree analysis was constructed with MEGA 5.2 software using the neighbor-joining method and the following parameters: bootstrap analysis of 1000 replicates, poisson model and pairwise deletion. The numbers at the nodes indicate the bootstrap values. SlDEAD30 and SlDEAD31 are marked with a black triangle and a red triangle, respectively. Accession numbers and corresponding references for the proteins used are as follows: Solanum lycopersicum: SlDEAD30, KJ739798; SlDEAD31, KJ713393. Arabidopsis thaliana: AtRH3, NM_001036866 [28]; AtRH9, NM_125492 [21]; AtRH22, NM_104691 [26]; AtRH36, NM_101494 [25]; AtRH52, NM_115719 [26]; STRS1, AY080680 [16]; STRS2, AY035114 [16]; LOS4, BT002444 [19]; RCF1, BT002030 [54]; AtCAF, AF187317 [27]; AtHEN2, AY050658 [55]. UPF1, AF484122 [56]. Oryza sativa: OsBIRH1, Q0DVX2 [17]; OsABP, LOC_Os06g33520 [57]; Hordeum vulgare: HVD1, AB164680 [58]; Glycine max: GmRH, FJ462142 [59]. Apocynum venetum: AvDH1, EU145588 [15].