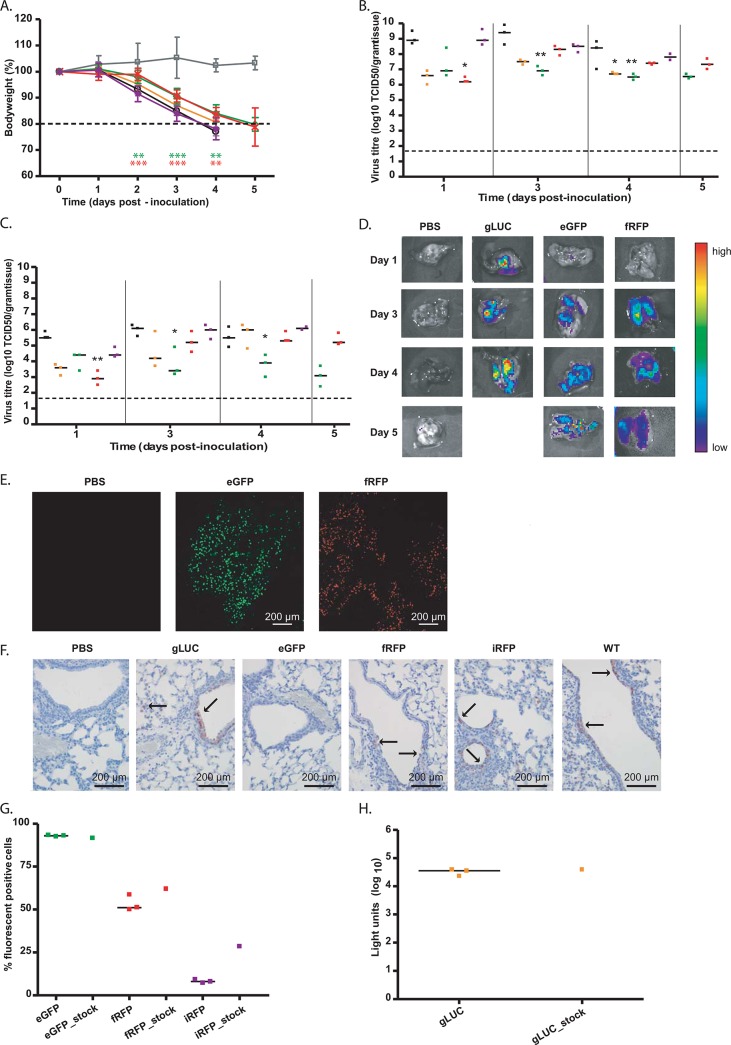
Fig 4. In vivo assessment of selected reporter viruses.
(A) Loss of bodyweight after intranasal inoculation of mice with PBS (grey square), WT (black circle), 2UP_PA_gLUC_sPR (yellow triangle), 2UP_PA_eGFP_sPR (green diamond), 2UP_PA_fRFP_sPR (red cross) and 2UP_PA_iRFP_dPR (purple square). Statistical significance was determined using two-way ANOVA with Bonferroni post-test and is depicted by two (p<0.01) or three (p<0.001) asterisks. Error bars indicate the standard error of the mean. Each group consisted of 6 mice for each time-point. Colours of the asterisk represent the different reporters (green for eGFP and red for fRFP virus). Lung (B) and nose (C) samples from 3 mice were homogenized and used for virus titrations at day 1, 3, 4 and 5. Black squares represent wild-type, orange squares gLUC, green squars eGFP, red squars fRFP and purple squares the iRFP virus. Statistical significance was determined using the Kruskal-Wallis and Dunn’s multiple comparison test and is depicted by one (p<0.05) or two (p<0.01) asterisks. Data are represented by x-y scatter plots showing individual virus titres, the bars represent the median and the dotted line the detection limit. (D) Lungs were imaged using the IVIS Spectrum. Colours represent the intensity of the signal with purple indicating the lowest and red the highest signal. (E) Confocal images of the entire lungs from mice inoculated with PBS, 2UP_PA_eGFP_sPR and 2UP_PA_fRFP_sPR virus. (F) Sections of lungs were stained for NP antigen. The red colour represents NP positive cells and arrows indicate examples of positive cells. (G) Lung homogenates and virus stocks were used to inoculated MDCK cells to determine the in vivo stability. The cells were stained for NP and within the influenza A positive population the percentage of fluorescent cells or luciferase activity (H) from the respective reporter was determined. Data are represented by x-y scatter plots showing individual virus titres, the bars represent the median and the dotted line the detection limit.