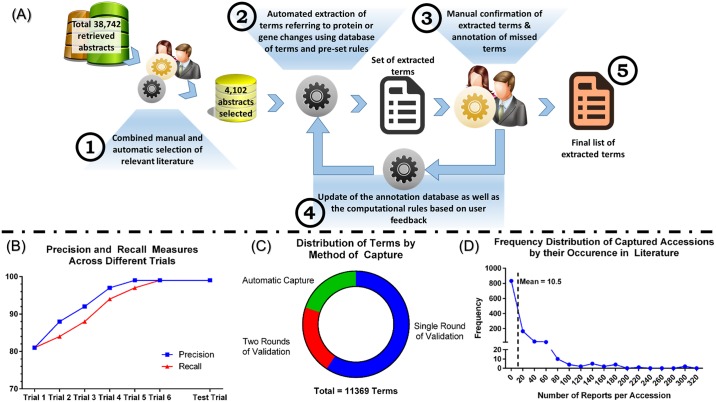
Fig 1. Design and output of the curation protocol.
(A) Overall Scheme of the Annotation and Term Detection Process. After articles are selected from PubMed using the search key, (1) a simultaneous manual and automatic selection of abstracts of papers referring to protein/gene changes after SCI to ensure best sensitivity and specificity of capture. Manual selection was performed by human annotators while automatic selection was performed through detection of desired terms in abstract (protein names, terms indicating changes in regulation, etc…). (2) We used an in-house tool that reads protein and genes names from online ontologies and then automatically detect exact or similar matches in the abstracts using pre-set rules. Then, (3) human annotators confirmed the extracted terms, delete false positives and (4) updated the tool with missed terms or rules for future use. Finally, (5) a full list of extracted terms is reported and referred to as the SCI meta-proteome (S3 Table). (B) Precision and recall results at different trials of annotation. Six trials of manual cross-validation and optimization were required to achieve more than 99.5% precision and recall of the tool (see methods for details). Test trial was performed to confirm the results. (C) Distribution of terms by method of capture. Around 60% of captured terms were validated by a single round of manual validation (by two human annotators), another 20% were validated by two rounds of manual validation due to low frequency or incomplete protein name description, and remaining 20% were automatically captured. (D) Frequency distribution of captured accessions by their literature occurrence.