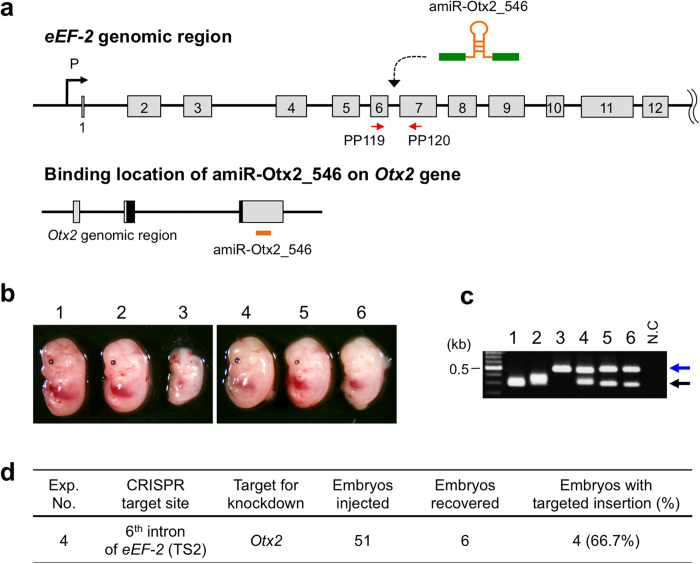
Figure 3. Targeted insertion of ssDNA encoding anti-Otx2 amiRNA by CRISPR/Cas9 system (Exp. 4).
(a) Schematics of targeted integration of amiR-Otx2_546 into intron 6 of eEF-2 gene (upper panel) and Otx2 genomic region showing amiR-Otx2_546 binding site (lower panel). The exons of Otx2 are shown as boxes and the boxes with homeodomain region are shaded black. Red arrows indicate the location of primer set (PP119/PP120) used for detection of fetuses with targeted insertion. (b) The resultant fetuses photographed at E14.5. (c) Genotyping of fetuses by PCR using primer set shown in (a). The embryo numbers in (b) and (c) correspond with each other. Expected fragment sizes: wild-type = 301-bp (black arrow), targeted insertion = 487-bp (blue arrow). (d) Targeted insertion efficiency. Fetuses containing functional amiRNA sequence were considered as ‘embryos with targeted insertions’ even though insertions were not fully accurate.