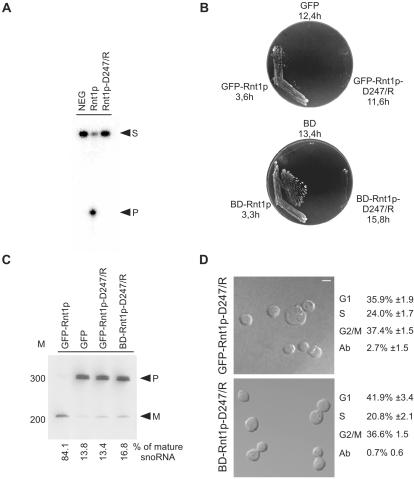
Figure 3.
Rnt1p catalytic activity is not required for cell division. A catalytically inactive mutant of Rnt1p (D247/R) was created by changing aspartic acid 247 to arginine. (A) In vitro cleavage assay of Rnt1p-D247/R. Mutant and wild-type recombinant proteins were tested for their ability to cleave, a 5′ end-labeled model of the Rnt1p natural substrate found at the 3′ end of U5 snRNA (16). NEG indicates RNA incubated under cleavage conditions in the absence of Rnt1p. Arrowheads indicate the position of the substrate (S) and the 5′ cleaved product (P). (B) Rnt1p catalytic activity is required for growth at 37°C. Δrnt1 cells expressing either GFP, or GFP fused with either Rnt1p or Rnt1p-D247/R (top), and cells expressing either BD or BD fused with either Rnt1p or Rnt1p-D247/R (bottom), were compared for growth on plates at 37°C. The doubling times of cells growing in liquid SCD media at 26°C are indicated under the name of each plasmid. (C) Northern blot analysis of RNA processing in vivo. RNA was extracted from the different yeast strains grown at 26°C, fractionated using denaturing PAGE, and transferred to nylon membrane. A probe specific to a known Rnt1p substrate (snoRNA, snR43) was used to visualize both mature (M) and premature RNAs (P). The bands were quantified using Instant Imager, and the ratios of mature to total snR43 were calculated (shown at bottom). The position of the 100-base pair DNA molecular weight marker is indicated on the left. (D) Phenotypic examination of cells expressing catalytically impaired Rnt1p. The phenotypes of Δrnt1 cells expressing either GFP-Rnt1p-D247/R, or Rnt1p in fusion with a heterologous nuclear localization signal (BDRnt1p-D247/R), were examined using confocal microscopy with the white colored bar representing 5 μm. Cells in different stages of the cell cycle were identified using bud size, and the cell number of an average of three independent experiments are indicated on the right. The cell cycle pattern of both cultures was also confirmed by FACS analysis (our unpublished data).