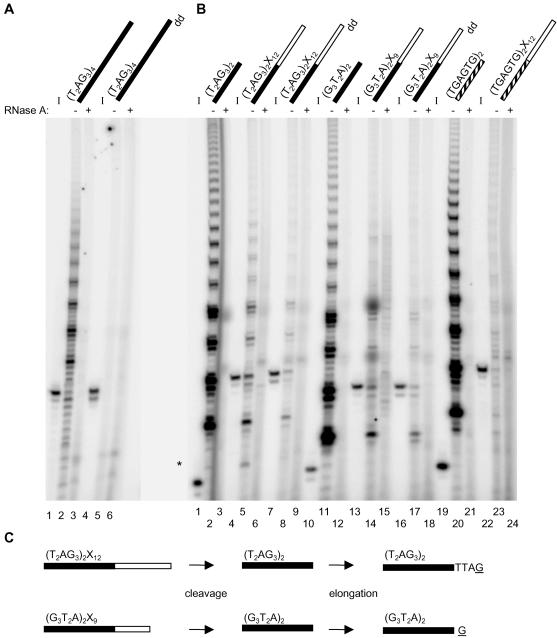
Figure 1.
(A) Telomerase elongation products generated by incubation with telomeric substrates. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and an oligonucleotide corresponding to four human telomeric repeats. Products of the standard elongation assay were electrophoresed on a 10% polyacrylamide denaturing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4). Oligonucleotides containing a dideoxynucleotide modification at the 3′ terminus are indicated by dd (lanes 4-6). Those samples pretreated with RNase A are indicated above the lane corresponding to their respective telomerase elongation reactions (+; lanes 3 and 6). (B) Telomerase elongation products generated by incubation with substrates containing 5′ telomeric DNA and 3′ nontelomeric DNA. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and the indicated oligonucleotide. Primer sequence composition is indicated by color code: black for telomeric DNA, white for nontelomeric DNA and striped for G-rich DNA. See Table 1 for oligonucleotide sequences. Products from the standard elongation assay were resolved on a 10% polyacrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, 10, 13, 16, 19, and 22). Oligonucleotides containing a dideoxynucleotide modification at the 3′ terminus are indicated by dd (lanes 7-9, 16-18). Perhaps due to an additional gel purification step, the dideoxynucleotide modified oligonucleotides generated slightly less intense elongation products relative to unmodified substrates (lanes 8 and 17, our unpublished results). Those samples pretreated with RNase A are indicated above the lane corresponding to their respective telomerase elongation reactions (+; lanes 3, 6, 9, 12, 15, 18, 21, and 24). An asterisk to the left indicates the position of the shortest product in lane 5. (C) Model for the cleavage and subsequent elongation of (T2AG3)2X12 and (G3T2A)2X9. Primer sequence composition is indicated by color: black for telomeric DNA and white for nontelomeric DNA. Primers are cleaved to expose telomeric DNA that becomes a substrate for elongation by telomerase (where G represents the incorporation of [α-32P]dGTP by telomerase). Note that regardless of whether cleavage occurs 5′ of or at the telomeric/nontelomeric DNA boundary, the elongation products will be similarly offset due to the permutation of telomeric DNA.