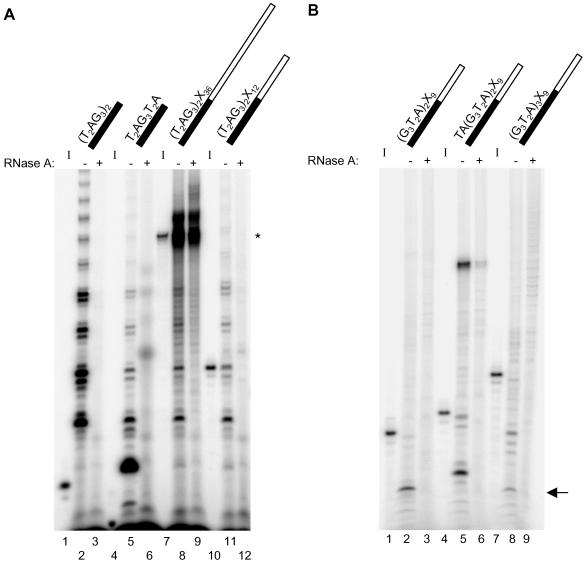
Figure 3.
Telomerase elongation products generated by incubation with chimeric oligonucleotides containing different lengths of 3′ nontelomeric or 5′ telomeric DNA. The standard elongation assay was conducted using partially purified human telomerase from Raji cells and the oligonucleotide indicated above each lane: black for telomeric DNA, white for nontelomeric DNA. Refer to Table 1 for oligonucleotide sequences. (A) Utilization of chimeric oligonucleotides with varying lengths of 3′ nontelomeric DNA. The standard elongation assay products were resolved on a 10% polyacrylamide sequencing gel. The 32P-5′ end-labeled input oligonucleotides are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, 7, and 10). RNase A pretreatment is indicated by + above the corresponding lane (lanes 3, 6, 9, and 12). Note that the products in the upper region of lane 8 were not RNase A sensitive (lane 9) and are therefore not telomerase dependent (see asterisk beside this region in the gel). (B) Utilization of chimeric oligonucleotides with varying lengths of 5′ telomeric DNA. Products generated in the standard elongation assay were electrophoresed on a 12% polyacrylamide denaturing gel. 32P-5′ end-labeled primers are shown to the left of their respective telomerase elongation reactions (I; lanes 1, 4, and 7). RNase A pretreatment is indicated by + above the corresponding lane (lanes 3, 6, and 9). The shortest major product generated by incubation with (G3T2A)2X9 and (G3T2A)3X9 is indicated by an arrow at right (lanes 2 and 8). Note that the products in the upper section of lanes 2, 5, and 8 were not sensitive to treatment with RNase A (lanes 3, 6, and 9, respectively) and are therefore not telomerase dependent.