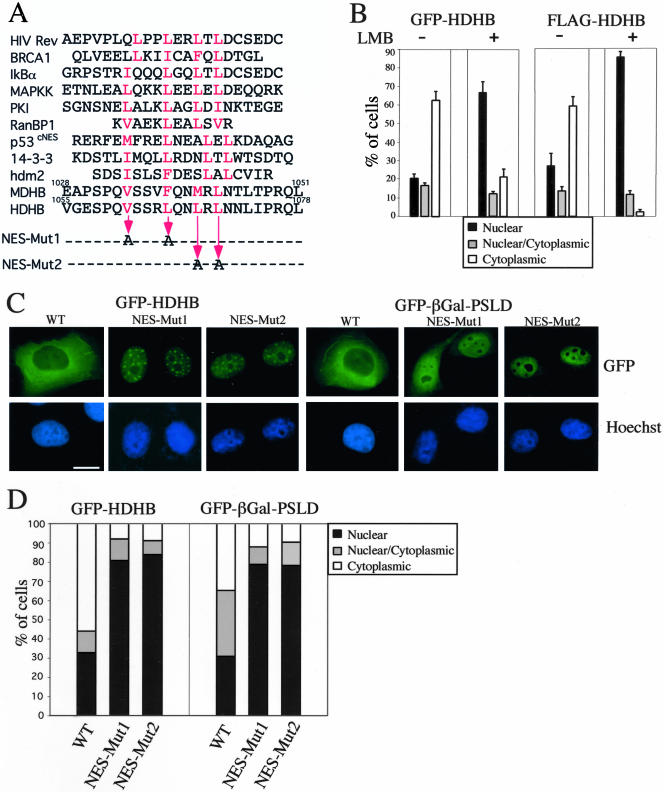
Figure 6.
Identification of a functional rev-type NES in SLD of HDHB. (A) Alignment of the putative NES in HDHB with those identified in other cell cycle-related proteins (Fabbro and Henderson, 2003). Superscripts above the amino acid sequence indicate residue numbers. Letters in red are the conserved aliphatic residues in the NES. Two pairs of residues in the putative NES in HDHB were mutated to alanine as indicated by the thin arrows to create Mut1 and Mut2. (B) GFP- and FLAG-tagged HDHB were transiently expressed in asynchronously growing U2OS cells with (+) or without (-) LMB to inhibit CRM1-mediated nuclear export. The subcellular localization of GFP-HDHB and FLAG-HDHB in asynchronous, G1, and S phase cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that sample. (C) GFP-HDHB and GFP-βGal-PSLD carrying the Mut1 or Mut2 mutations, or the corresponding proteins without the mutations, were transiently expressed in asynchronous U2OS cells and visualized by fluorescence microscopy. Cells showing the most frequently observed fluorescence pattern are shown. Nuclei were stained with Hoechst dye. Bar, 10 μm. (D) The subcellular localization of wild-type and mutant GFP-HDHB and GFP-βGal-PSLD in asynchronous U2OS cells was quantified and expressed as a percentage of the total number of GFP-positive cells in that sample.