Abstract
Purpose: To investigate the significance of mammalian target of rapamycin (mTOR) in colorectal cancers. mTOR has recently been suggested as a prognostic biomarker and therapeutic target in an array of human cancers. Findings: phospho-mTOR (p-mTOR) expression was analyzed by immunohistochemistry (IHC) on a tissue microarray containing 1800 colorectal cancers (CRC). Clinical follow-up data were available from all cancer patients. Positive p-mTOR immunostaining was seen in 83.5% of 1640 interpretable CRC and was considered weak in 862 (52.5%) and strong in 508 cases (31.0%). Matching clinico-pathological parameters were available in 1580 cases. p-mTOR staining was more frequent in tubular adenocarcinomas than in the less common histological subtypes (mucinous, medullary, signet cell; P=0.0163) and significantly linked to carcinomas of the left-sided colon and rectum as compared to right-sided CRC (P=0.0066). There was no significant association between p-mTOR expression and patients’ gender, tumor stage, tumor grade or nodal status. In a survival analysis, p-mTOR IHC status of all CRC was unrelated to patient survival (P=0.702). In a multivariate analysis including pT, pN, tumor grade, tumor localization and p-mTOR expression, only pT, pN (both P<0.0001) and grade (P=0.0001) showed prognostic impact, but not tumor localization (P=0.9472) or p-mTOR expression (P=0.8879). Conclusion: Our observations indicate that p-mTOR overexpression is abundant in CRC and linked to left-sided tumor localization. The high frequency and overexpression of p-mTOR is providing further rationale for targeting this pathway therapeutically in CRC patients. However, a prognostic role of p-mTOR overexpression in CRC could not be confirmed.
Keywords: Colorectal cancer, phospho-mTOR, tissue microarray, immunohistochemistry
Introduction
Colorectal cancer (CRC) is the fourth most common malignant disease with over one million novel cases and over 500.000 deaths each year worldwide [1]. Although recent advances in the management of the disease have improved outcomes, CRC remains the second leading cause of cancer-related death in Western countries [1]. In advanced metastatic colorectal cancer (mCRC), surgery alone is not curative and therefore adjuvant chemotherapy is needed. New anticancer drugs have improved the standard chemotherapy treatment of CRC and there is much promise in molecular-targeted therapy.
Mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase that belongs to the phosphoinositide 3-kinase (PI3 K)-related kinase family. It constitutes the core of an evolutionarily conserved pathway that regulates cell growth and proliferation in normal human cells [2,3]. Moreover, mTOR signaling activity is associated with cancer cell growth and survival [4,5]. Abnormal mTOR expression has been described in a variety of human tumors [6-8] and was found to be associated with poor prognosis [9-11]. In CRC, mTOR overexpression is likely to be involved in the development and progression of the disease and is linked to cancer initiation, invasiveness, and progression [12]. Recently, inhibitors of proteins that are involved in mTOR signaling have been under active preclinical or clinical investigation for cancer therapy [13-19].
To further expand our knowledge on the relevance of (phospho-) mTOR (p-mTOR) expression in CRC, we analyzed a tissue microarray including a series of 1800 cancers with clinical follow up and extensive molecular data. Since mTor phosphorylated at Ser 2448 (p-mTOR Ser 2448) is the activated variant of mTOR, we performed p-mTOR immunostaining on our TMA and evaluated the correlation of the activated protein with clinico-pathological parameters.
Material and methods
Patients and tissue microarray (TMA) construction
Two different TMAs with a total of 1800 CRC samples were included in this study. The first TMA was manufactured from resection specimens of 1420 CRC patients at the Institute of Pathology of the University Hospital of Basel). None of the patients received neo-adjuvant or adjuvant therapy. Raw survival data were obtained from the responsible physicians for all of the 1420 patients. The median follow up time was 46 months (range 1-152 months). The second TMA included samples from 380 CRC patients, whose tumor resection specimens were examined at the Institute of Pathology of the University Medical Center, Hamburg-Eppendorf. Also for this TMA, raw survival data were available for all of the 380 patients with a median follow up period of 36 months (range 1-179 months). TMA construction was as described [20]. In brief, hematoxylin and eosin-stained sections were made from each block to define representative tumor regions. Tissue cylinders with a diameter of 0.6 mm were then punched from tumor areas of each “donor” tissue block using a home-made semi-automated precision instrument and brought into empty recipient paraffin blocks. Four µm sections of the resulting TMA blocks were transferred to an adhesive coated slide system (Instrumedics Inc., Hackensack, New Jersey). Patient information and clinical data such as age, sex, localization and type of the tumor, pTNM-stage and carcinoma grade were retrospectively retrieved from clinical and pathological databases (Table 1). All tumors were re-classified by two pathologists (PS, AM). Follow-up data were obtained from local cancer register boards or via attending physicians. For statistical analyses, tumor localizations were grouped as follows: right-sided cancer (cecum, ascending colon), cancer of the transverse colon, cancer of the left-sided colon (descending colon, sigmoid colon) and rectum. The utilization of tissues and clinical data was according to the Hamburger Krankenhaus Gesetz (§12 HmbKHG) and approved by our local Ethical Committee.
Table 1.
Hospho-mTOR (p-mTOR) immunohistochemistry and clinico-pathological features
| p-mTOR | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Negative | Weak | Strong | ||||
|
|
||||||
| N | % | % | % | P value | ||
| Gender | Male | 794 | 17.4 | 49.9 | 32.7 | |
| Female | 786 | 15.2 | 55.0 | 29.8 | 0.1225 | |
| Tumor grade | G1 | 25 | 8.0 | 44.0 | 48.0 | |
| G2 | 1393 | 15.9 | 52.9 | 31.2 | ||
| G3 | 162 | 22.2 | 49.4 | 28.4 | 0.1050 | |
| Tumor stage | Pt1 | 68 | 8.8 | 50.0 | 41.2 | |
| Pt2 | 255 | 15.3 | 52.9 | 31.8 | ||
| Pt3 | 1019 | 16.4 | 52.1 | 31.5 | ||
| Pt4 | 238 | 19.7 | 53.8 | 26.5 | 0.1972 | |
| Nodal status | Pn0 | 816 | 15.8 | 54.0 | 30.2 | |
| Pn1 | 428 | 16.4 | 49.8 | 33.9 | ||
| Pn2 | 336 | 17.9 | 51.8 | 30.3 | 0.5672 | |
| Tumor type | Tubular | 1526 | 15.9 | 52.2 | 31.9 | |
| Others | 54 | 30.8 | 57.7 | 11.5 | 0.0163 | |
| Localization | Coecum, ascending | 383 | 19.1 | 51.4 | 29.5 | |
| Transverse | 137 | 27.7 | 41.6 | 30.7 | ||
| Descending | 67 | 19.4 | 49.5 | 31.3 | ||
| Sigmoid | 410 | 15.1 | 54.4 | 30.5 | ||
| Rectum | 582 | 12.5 | 54.5 | 33.0 | 0.0066 | |
| Total | 1580 | |||||
Immunohistochemistry
Freshly cut TMA sections were immunostained on one day and in one experiment. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121°C in pH 2.0 in target retrieval solution (Biogenex, San Ramon, USA). Primary antibody specific for mTOR phosphorylated at Ser2448 (rabbit polyclonal phospho-mTOR antibody Ser2448; Cell signaling technology, Germany, Frankfurt am Main; dilution 1:150) was applied at 37°C for 60 minutes. Bound antibody was then visualized using the EnVision Kit (Dako, Glostrup, Denmark) according to the manufacturer’s directions. Only cytoplasmic staining was considered. The staining results were categorized into three groups. Tumors without any staining were considered p-mTOR “negative”. Tumors with 1+ or 2+ staining in up to 50% of cells or 3+ staining in up to 20% of cells were considered “weakly positive”. Tumors with 2+ staining in >50% or 3+ staining in >20% were considered “strongly positive”. This categorization was also used in an earlier study of our group [21].
Statistics
Statistical calculations were performed with JMP® 10.0.2 software (2012 SAS Institute Inc., NC, USA). Contingency tables and the chi²-test were performed to search for associations between molecular parameters and tumor phenotype. Survival curves were calculated according to Kaplan-Meier. The Log-Rank test was applied to detect significant survival differences between groups. Cox proportional hazards regression analysis was performed to test the statistical independence and significance between pathological and clinical variables.
Results
Phospho-mTOR-immunohistochemistry
A total of 116 of 1800 tissue samples (6.4%) were non-informative due to either absence of unequivocal cancer tissue, uninterpretable staining or complete loss of tissue. Normal colorectal epithelial cells typically did not show any staining for p-mTOR. Positive immunostaining was seen in 83.5% of 1640 interpretable CRC. Immunostaining was typically cytoplasmic. Representative examples of p-mTOR stained cancers are shown in Figure 1. p-mTOR staining was considered weak in 862 (52.5%) and strong in 508 cases (31.0%). Clinico-pathological parameters in association with p-mTOR IHC results were available for 1580 cases. The relationship between p-mTOR staining, tumor phenotype and clinical parameters is shown in Table 1. p-mTOR staining was more frequent in tubular adenocarcinomas than in the less common histological subtypes (mucinous, medullary, signet cell; P=0.0163, Table 1). Moreover, p-mTOR expression levels were significantly related to the tumor localization. p-mTOR expression was more frequent in carcinomas of the sigmoid colon (84.9%) and rectum (87.5%) as compared to right-sided CRC (80.9%, Table 1; Figure 2; P=0.0066). There was no significant association between p-mTOR expression and patients’ gender, tumor stage, tumor grade or nodal status (Table 1).
Figure 1.
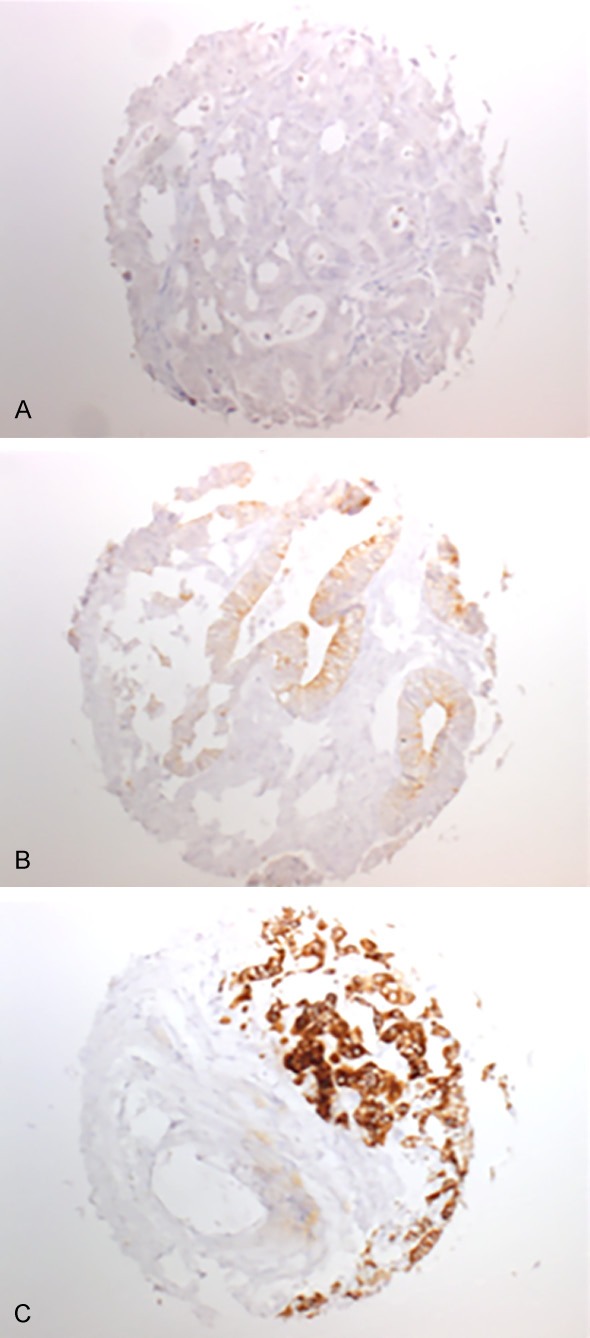
TMA samples of colorectal cancers showing negative (A), weak (B) and strong (C) immunostaining for p-mTOR (magnification 50×).
Figure 2.
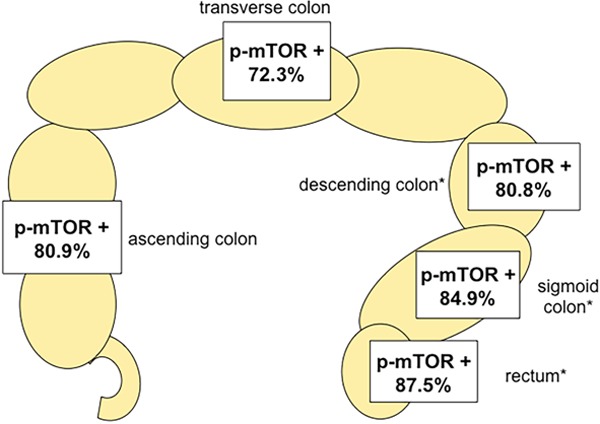
Positive p-mTOR immunostaining (weak and strong) is significantly related to left-sided tumor localization, *P=0.0066.
Survival analysis
As expected, high tumor grade and stage as well as advanced nodal status were associated with poor patient survival (Figure 3A-C; P<0.0001 each) while histological tumor type was unrelated to clinical outcome (P=0.6279, Figure not shown). Left-sided CRC (distal to the splenic flexure) was associated with a better prognosis (P=0.0161; Figure 3D) as compared to cancers of the right-sided colon. p-mTOR IHC status of all CRC was unrelated to patient survival (P=0.7020; Figure 3E). These associations also held true in the subset of tubular carcinomas only (data not shown).
Figure 3.
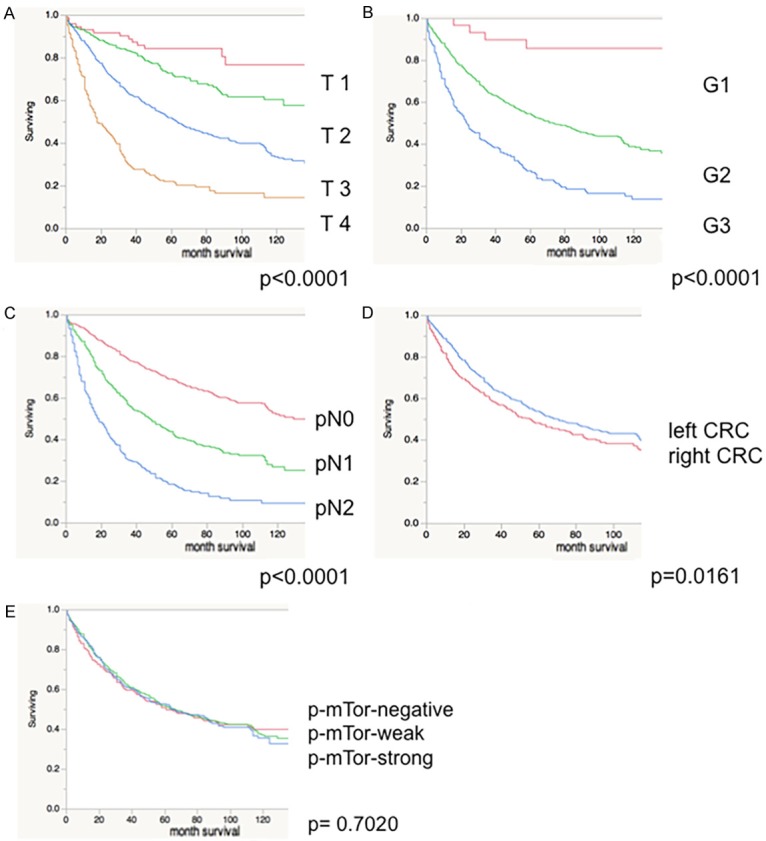
Patient survival is related to tumor stage (A), tumor grade (B), nodal status (C) and tumor localization (D), but not to p-mTOR IHC (E; Red-negative; Green- weak; Blue-strong p-mTOR IHC).
Multivariate analysis
In a multivariate analysis including pT, pN, tumor grade, tumor localization and p-mTOR expression only pT, pN (both P<0.0001) and grade (P=0.0001) showed prognostic impact, but not tumor localization (P=0.9472) or p-mTOR expression (P=0.8879; Table 2).
Table 2.
Multivariate analysis of phospho-mTOR expression in colorectal cancer
| N Analyzable | P-value | ||||
|---|---|---|---|---|---|
|
| |||||
| pT | pN | Grading | Tumour localization | p-mTOR expression | |
| 1580 | <0.0001 | <0.0001 | 0.0001 | 0.9472 | 0.8879 |
Discussion
The results of this study show that p-mTOR (Ser2448) is abundant in CRC (83.8%) and that it is associated with left-sided tumor localization.
The high frequency of p-mTOR found in this study is comparable to previous results. Wang et al. described about 61% p-mTOR positive CRC [22]. Interestingly, Cai et al. had previously reported that p-mTOR overexpression is predictive of poor outcome in stage II CRC [23]. In contrast, we could not find a significant association between p-mTOR expression and prognosis, despite other features, such as tumor grade and tumor stage, being linked to outcome in this patient cohort. This observation held true when only stage II CRC and p-mTOR expression was examined (data not shown). However, the differences in p-mTOR expression between tumors of different grade and stage were quite small in terms of absolute numbers. The strong significance seems to be caused by the high number of analyzed cases in our study (n=1580) providing strong statistical power. The strong association between classical prognostic features such as tumor stage, tumor grade and nodal status and prognosis in our patent set provide indirect proof for the validity of our clinical data. Therefore, we believe that our findings argue against a clinical utility of p-mTOR expression as a prognostic biomarker in colorectal cancer patients.
Interestingly, p-mTOR expression levels were significantly higher in left-sided than in right-sided CRC (Table 2; Figure 2). This finding obtains support in a study by Alqurashi et al showing that high levels of mTOR RNA were found more frequently in left-sided CRC [12]. The reason for this observation is unclear. It could be hypothesized that pathways leading to increased p-mTOR expression are less often activated in patients with hereditary-non-polyposis colon cancer syndromes (HNPCC) or sporadic colorectal cancers exhibiting microsatellite instability (MSI), which are more often localized in the right colon. This observation does not rule out other causes for the left colon preference of p-mTOR. CRCs that arise on the left or right side exhibit substantial differences in gene expression and signal transduction patterns. In fact, it is expected that every alteration occurring more frequently in the left colon than in the right colon should be statistically associated with lower MSI frequency, as MSI is known to preferably occur in the right colon [24].
Based on the high frequency of p-mTOR positivity seen in this study and the concordance of our results with previous data, it appears unlikely, that our TMA-based approach, analyzing only limited amounts of tumor tissue per patient (1 spot of 0.6mm diameter) has led to a significant number of false negative cases.
The high frequency and overexpression of p-mTOR provides further rationale for targeting this pathway therapeutically in CRC patients. It has been reported that concomitant BRAF and PI3K/mTOR blockade is required for effective treatment of BRAF (V600E) colorectal cancer [25]. Recently, it has been suggested that combinations of drugs targeting BRAF (and/or MEK) and eIF4F-a downstream complex of the PI (3)K-AKT-mTOR pathway-may overcome most of the resistance mechanisms arising in BRAF (V600E)-mutant cancers, including CRC [26]. Positive results with mTOR antagonists in renal carcinoma and several lymphoma subtypes have recently led to the initiation of a variety of new clinical trials with other types of tumors, including gastric, endometrial, and non-small cell lung cancer as well as sarcoma and neuroendocrine tumors [27]. Regarding CRC, the treatment with everolimus, an oral inhibitor of mTOR, showed efficacy in patients with metastatic colorectal cancers in phase I studies [28,29]. In a phase II study, among patients receiving everolimus, those with a KRAS mutation experienced significantly shorter median overall survival compared with those with wild-type KRAS [30]. In another phase II study, the oral combination of tivozanib and everolimus was well tolerated, with stable disease achieved in 50% of patients with refractory, metastatic colorectal cancer [31]. Future studies may be needed evaluating everolimus in combination with other agents or in patients with dysregulation of the PI3K/Akt/mTOR pathway.
Although the presence of a target on tumor cells does not guarantee successful therapy by itself, our data at least indicate that mTOR might play a functional role in CRC and could potentially identify patients benefitting from therapy with mTOR antagonists.
In summary, our data show that p-mTOR expression occurs frequently in CRC with a clear preference for left-sided tumors. The high frequency and overexpression of p-mTOR provides further rationale for targeting this pathway therapeutically in CRC patients. Despite a relationship between p-mTOR expression and unfavorable tumor phenotype, previous reports on a possible prognostic role of p-mTOR in CRC could not be confirmed.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Tsang CK, Zheng XF. TOR-in(g) the nucleus. Cell Cycle. 2007;6:25–29. doi: 10.4161/cc.6.1.3675. [DOI] [PubMed] [Google Scholar]
- 4.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 6.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 7.Bellacosa A, de Feo D, Godwin AK, Bell DW, Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, Ferrandina G, Benedetti Panici P, Mancuso S, Neri G, Testa JR. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 8.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 9.Pantuck AJ, Seligson DB, Klatte T, Yu H, Leppert JT, Moore L, O’Toole T, Gibbons J, Belldegrun AS, Figlin RA. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- 10.Bunkholt Elstrand M, Dong HP, Odegaard E, Holth A, Elloul S, Reich R, Trope CG, Davidson B. Mammalian target of rapamycin is a biomarker of poor survival in metastatic serous ovarian carcinoma. Hum Pathol. 2010;41:794–804. doi: 10.1016/j.humpath.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Huang Y, Li J, Wang Z. The mTOR pathway is associated with the poor prognosis of human hepatocellular carcinoma. Med Oncol. 2010;27:255–261. doi: 10.1007/s12032-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 12.Alqurashi N, Gopalan V, Smith RA, Lam AK. Clinical impacts of mammalian target of rapamycin expression in human colorectal cancers. Hum Pathol. 2013;44:2089–2096. doi: 10.1016/j.humpath.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Gulhati P, Cai Q, Li J, Liu J, Rychahou PG, Qiu S, Lee EY, Silva SR, Bowen KA, Gao T, Evers BM. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuge R, Kitadai Y, Shinagawa K, Onoyama M, Tanaka S, Yasui W, Chayama K. mTOR and PDGF Pathway Blockade Inhibits Liver Metastasis of Colorectal Cancer by Modulating the Tumor Microenvironment. Am J Pathol. 2015;185:399–408. doi: 10.1016/j.ajpath.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Brana I, Berger R, Golan T, Haluska P, Edenfield J, Fiorica J, Stephenson J, Martin LP, Westin S, Hanjani P, Jones MB, Almhanna K, Wenham RM, Sullivan DM, Dalton WS, Gunchenko A, Cheng JD, Siu LL, Gray JE. A parallel-arm phase I trial of the humanised anti-IGF-1R antibody dalotuzumab in combination with the AKT inhibitor MK-2206, the mTOR inhibitor ridaforolimus, or the NOTCH inhibitor MK-0752, in patients with advanced solid tumours. Br J Cancer. 2014;111:1932–1944. doi: 10.1038/bjc.2014.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciuffreda L, Di Sanza C, Incani UC, Milella M. The mTOR Pathway: A New Target in Cancer Therapy. Curr Cancer Drug Targets. 2010;10:484–495. doi: 10.2174/156800910791517172. [DOI] [PubMed] [Google Scholar]
- 17.Thiery-Vuillemin A, Theodore C, Jacobasch L, Schmitz J, Papandreou C, Guillot A, Emmanouilides C, Slimane K, Kelkouli N, Kim S, Nguyen Tan Hon T. Efficacy and Safety of Sequential Use of Everolimus in Patients With Metastatic Renal Cell Carcinoma Previously Treated With Bevacizumab With or Without Interferon Therapy: Results From the European AVATOR Study. Clin Genitourin Cancer. 2015;13:231–8. doi: 10.1016/j.clgc.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Tsimafeyeu I, Snegovoy A, Varlamov S, Safina S, Varlamov I, Gurina L, Manzuk L. Everolimus in patients with metastatic renal cell carcinoma previously treated with bevacizumab: a prospective multicenter study CRAD001LRU02T. Target Oncol. 2014 doi: 10.1007/s11523-014-0347-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Courtney KD, Manola JB, Elfiky AA, Ross R, Oh WK, Yap JT, Van den Abbeele AD, Ryan CW, Beer TM, Loda M, Priolo C, Kantoff P, Taplin ME. A Phase I Study of Everolimus and Docetaxel in Patients With Castration-Resistant Prostate Cancer. Clin Genitourin Cancer. 2015;13:113–23. doi: 10.1016/j.clgc.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 21.Marx A, Simon P, Simon R, Mirlacher M, Izbicki JR, Yekebas E, Kaifi JT, Terracciano L, Sauter G. AMACR expression in colorectal cancer is associated with left-sided tumor localization. Virchows Arch. 2008;453:243–248. doi: 10.1007/s00428-008-0646-1. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Chen J, Guo F, Chen H, Duan Z, Wei MY, Xu QM, Wang LH, Zhong MZ. Clinical significance of mTOR and p-mTOR protein expression in human colorectal carcinomas. Asian Pac J Cancer Prev. 2011;12:2581–2584. [PubMed] [Google Scholar]
- 23.Cai Z, Ke J, He X, Yuan R, Chen Y, Wu X, Wang L, Wang J, Lan P, Wu X. Significance of mTOR signaling and its inhibitor against cancer stem-like cells in colorectal cancer. Ann Surg Oncol. 2014;21:179–188. doi: 10.1245/s10434-013-3146-8. [DOI] [PubMed] [Google Scholar]
- 24.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 25.Coffee EM, Faber AC, Roper J, Sinnamon MJ, Goel G, Keung L, Wang WV, Vecchione L, de Vriendt V, Weinstein BJ, Bronson RT, Tejpar S, Xavier RJ, Engelman JA, Martin ES, Hung KE. Concomitant BRAF and PI3K/mTOR blockade is required for effective treatment of BRAF(V600E) colorectal cancer. Clin Cancer Res. 2013;19:2688–2698. doi: 10.1158/1078-0432.CCR-12-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F, Mateus C, Routier E, Kamsu-Kom N, Agoussi S, Eggermont AM, Desaubry L, Robert C, Vagner S. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513:105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 27.Yuan R, Kay A, Berg WJ, Lebwohl D. Targeting tumorigenesis: development and use of mTOR inhibitors in cancer therapy. J Hematol Oncol. 2009;2:45. doi: 10.1186/1756-8722-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, Martinelli E, Ramon y Cajal S, Jones S, Vidal L, Shand N, Macarulla T, Ramos FJ, Dimitrijevic S, Zoellner U, Tang P, Stumm M, Lane HA, Lebwohl D, Baselga J. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J. Clin. Oncol. 2008;26:1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 29.O’Donnell A, Faivre S, Burris HA 3rd, Rea D, Papadimitrakopoulou V, Shand N, Lane HA, Hazell K, Zoellner U, Kovarik JM, Brock C, Jones S, Raymond E, Judson I. Phase I pharmacokinetic and pharmacodynamic study of the oral mammalian target of rapamycin inhibitor everolimus in patients with advanced solid tumors. J. Clin. Oncol. 2008;26:1588–1595. doi: 10.1200/JCO.2007.14.0988. [DOI] [PubMed] [Google Scholar]
- 30.Ng K, Tabernero J, Hwang J, Bajetta E, Sharma S, Del Prete SA, Arrowsmith ER, Ryan DP, Sedova M, Jin J, Malek K, Fuchs CS. Phase II study of everolimus in patients with metastatic colorectal adenocarcinoma previously treated with bevacizumab-, fluoropyrimidine-, oxaliplatin-, and irinotecan-based regimens. Clin Cancer Res. 2013;19:3987–3995. doi: 10.1158/1078-0432.CCR-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolpin BM, Ng K, Zhu AX, Abrams T, Enzinger PC, McCleary NJ, Schrag D, Kwak EL, Allen JN, Bhargava P, Chan JA, Goessling W, Blaszkowsky LS, Supko JG, Elliot M, Sato K, Regan E, Meyerhardt JA, Fuchs CS. Multicenter phase II study of tivozanib (AV-951) and everolimus (RAD001) for patients with refractory, metastatic colorectal cancer. Oncologist. 2013;18:377–378. doi: 10.1634/theoncologist.2012-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]


