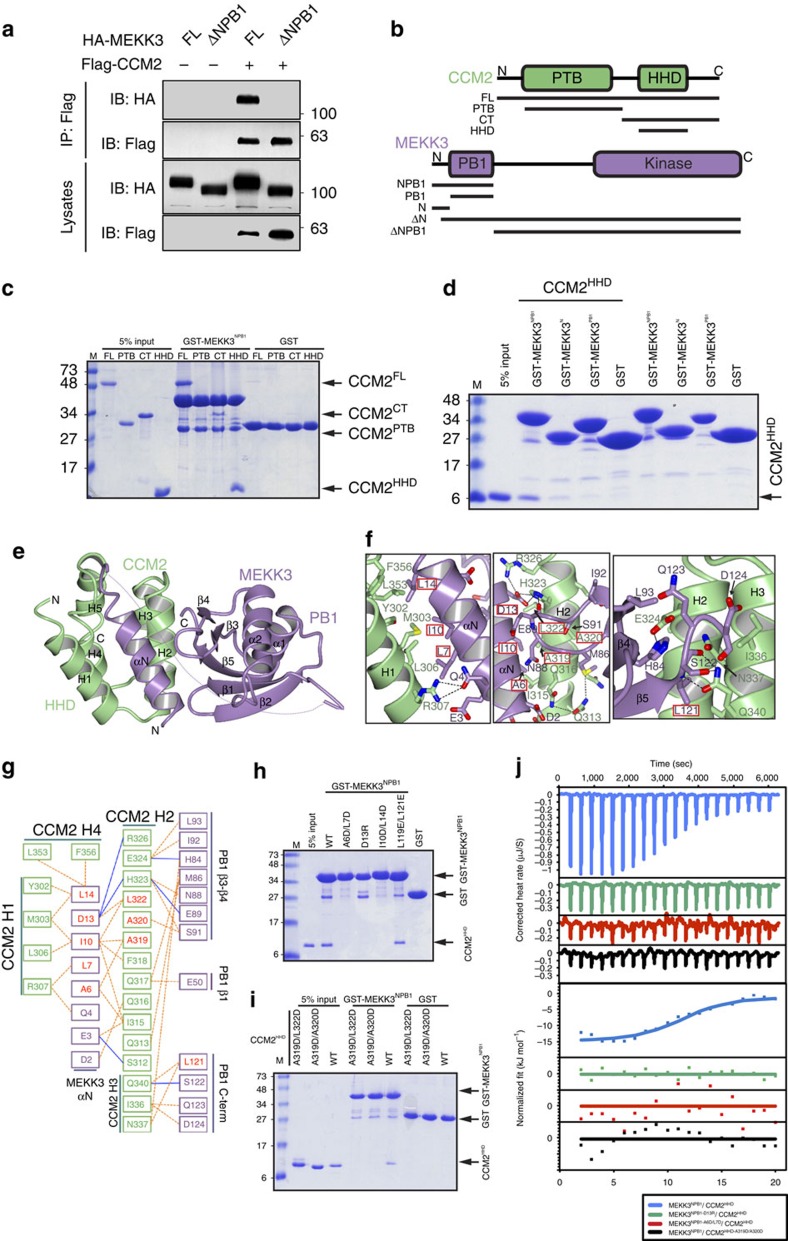
Figure 2. MEKK3 uses its NPB1 region to interact with the HHD domain of CCM2.
(a) Immunoprecipitation (IP) of Flag–CCM2 after cotransfection with HA–MEKK3FL or HA–MEKK3ΔNPB1 into 293T cells. Whole-cell lysates and immunoprecipitates were analysed by immunoblotting (IB) with the indicated antibodies. (b) Domain schematic of CCM2 (top) and MEKK3 (bottom). Black lines indicate constructs used in this study. Domains are indicated as PTB (Phosphotyrosine Binding), HHD (Harmonin homology domain), PB1 (Phox/Bem1p) and kinase. N and C termini are indicated. (c) Pull-down assay. GST–MEKK3NPB1 was immobilized on glutathione Sepharose beads and tested for ability to pull-down purified regions of CCM2. Coomassie stained SDS–PAGE analysis of washed beads. (d) Pull-down assay. Immobilized GST–MEKK3NPB1, GST–MEKK3N or GST–MEKK3PB1 were incubated with purified CCM2HHD. Coomassie-stained SDS–PAGE of washed beads. (e) Cocrystal structure of MEKK3NPB1 in complex with CCM2HHD. CCM2HHD is shown in green and MEKK3NPB1 in purple. Secondary structure elements are labelled. Disordered residues indicated by a dotted line. N and C termini are indicated. (f) Close-up views of the main interaction sites between MEKK3NPB1 and CCM2HHD. Left, CCM2 helix H1 with MEKK3 helix αN. Centre, CCM2 helix H2 with MEKK3 helix αN and PB1 domain. Right, CCM2 helices H2 and H3 with MEKK3 PB1 domain. Hydrogen-bonds shown as dotted lines. Residues mutated in this study are boxed in red. Structural figures generated using CCP4mg36. (g) Map of MEKK3NPB1:CCM2HHD interactions. MEKK3 residues are shown in purple and CCM2 residues in green. Residues mutated in this study are shown in red. Dotted lines indicate hydrophobic interactions and solid lines ionic interactions. Interactions defined by PDBSum37. (h) Pull-down assay. Crystallographically determined mutations to GST–MEKK3NPB1 interrupts pull-down of wild-type CCM2HHD. MEKK3 mutants, A6D/L7D, D13R, I10D/L14D and L119E/L121E are indicated. Coomassie stained SDS–PAGE of washed beads. (i) Pull-down assay. Introduction of crystallographically determined mutations to CCM2HHD interrupts pull-down with wild-type GST–MEKK3NPB1. CCM2 mutants A319D/L322D and A319D/A320D are indicated. Coomassie stained SDS–PAGE of washed beads. (j) Isothermal titration calorimetry. CCM2HHD was titrated into MEKK3NPB1. Crystallographically defined mutants tested for MEKK3 were D13R and A6D/L7D and for CCM2 was A319D/A320D38.