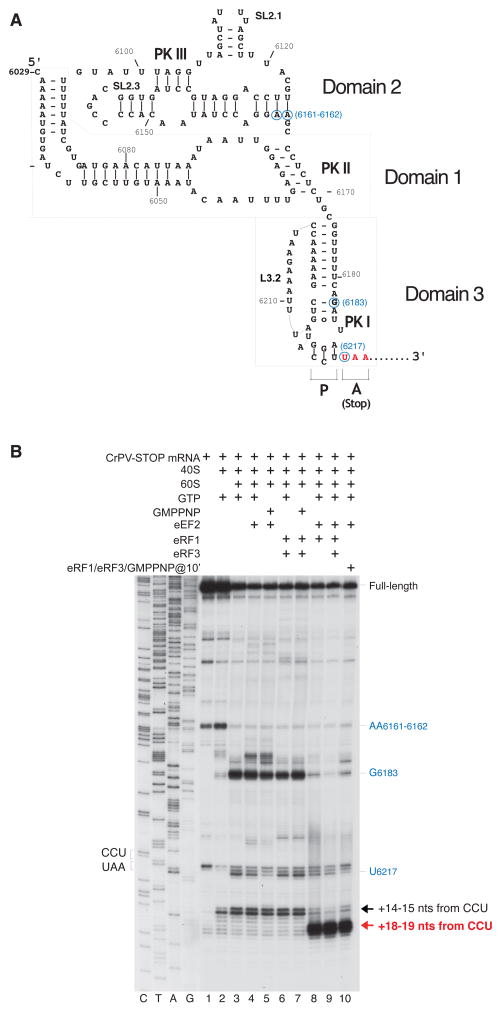
Figure 1. eEF2-dependent association of eRF1 and eRF1/eRF3 with 80S ribosomal complexes assembled on CrPV-STOP mRNA.
(A) Secondary structure of the CrPV IRES with the first Ala codon mutated to an UAA stop codon (in red). Blue circles indicate the positions of toe-prints (panel B) caused by the contacts of the IRES with the 40S and 60S ribosomal subunits. (B) Toe-printing analysis of binding of eRF1 and eRF3 to 80S ribosomal complexes assembled on CrPV-STOP mRNA, depending on the presence of eEF2. Toe-prints corresponding to the pre-translocated ribosomal complexes (+14–15 nts from the CCU codon) are indicated by a black arrow. Toe-prints corresponding to the eRF1- or eRF1/eRF3-associated post-translocated ribosomal complexes (+18–19 nts from the CCU codon) are indicated by a red arrow. The +4 nt toe-print shift in post-translocated complexes includes the +2nt shift due to the presence of eRF1 (Alkalaeva et al., 2006). Additional toe-prints caused by the contacts of the IRES with the 40S subunit (at AA6161–6162) and with the 60S subunit (U6217 and G6183) are consistent with previous reports (Wilson et al., 2000b; Hellen and Pestova, 2003) and are shown in blue.