Abstract
The goal of this study was to characterize the molecular mechanisms underlying cetuximab-mediated upregulation of HLA class I antigen-processing machinery components in head and neck cancer (HNC) cells and to determine the clinical significance of these changes in cetuximab-treated HNC patients. Flow cytometry, signaling studies and chromatin immunoprecipitation (ChIP) assays were performed using HNC cells treated with cetuximab alone or with Fcγ receptor (FcγR)-bearing lymphocytes to establish the mechanism of EGFR-dependent regulation of HLA APM expression. A prospective phase II clinical trial of neoadjuvant cetuximab was utilized to correlate HLA class I expression with clinical response in HNC patients. EGFR blockade triggered STAT1 activation and HLA upregulation, in a src homology-containing protein (SHP)-2-dependent fashion, more prominently in HLA-B/C than in HLA-A alleles. EGFR signaling blockade also enhanced IFNγ receptor 1 (IFNAR) expression, augmenting induction of HLA class I and TAP1/2 expression by IFNγ, which was abrogated in STAT1−/− cells. Cetuximab enhanced HNC cell recognition by EGFR853–861-specific CTLs, and notably enhanced surface presentation of a non-EGFR peptide (MAGE-3271–279). HLA class I upregulation was significantly associated with clinical response in cetuximab-treated HNC patients. EGFR induces HLA downregulation through SHP-2/STAT1 suppression. Reversal of HLA class I downregulation was more prominent in clinical responders to cetuximab therapy, supporting an important role for adaptive immunity in cetuximab antitumor activity. Abrogating EGFR-induced immune escape mechanisms and restoring STAT1 signaling to reverse HLA downregulation using cetuximab should be combined with strategies to enhance adaptive cellular immunity.
Keywords: Cetuximab, immune escape, EGFR, cytotoxic T cells, ADCC, head and neck cancer, immunotherapy
Introduction
The mitogenic activity of the epidermal growth factor receptor (EGFR) has provided the rationale for the development of inhibitory strategies to block EGFR signaling, using tyrosine kinase inhibitors (TKI) and EGFR-targeted monoclonal antibodies (mAb). This strategy has been shown to be effective, since the EGFR-specific mAb cetuximab has been approved by the FDA for head and neck cancer (HNC) and colorectal cancer (1), and yet no biomarker of clinical activity has been determined. EGFR signaling also influences the expression of immunologically relevant molecules in HNC cells, including STAT1-mediated HLA and antigen-processing machinery (APM) components (2–4), implying an important impact on adaptive immunity due to EGFR overexpression. However the precise mechanism, functional effect(s), and clinical significance of these findings have not yet been determined.
Mechanism(s) of HLA class I APM component deficiency are still not clear, despite the importance of avoidance of lysis by cytotoxic T lymphocytes (CTL) (5–9). Overexpression of EGFR, its ligands and concomitant downstream signaling facilitates HNC proliferation by activating multiple pathways (10). Previously, we demonstrated the reversal of HLA class I and APM component deficiency in HNC using the STAT1 agonist IFNγ, which enhanced CTL-mediated lysis and induced a higher level of peptide:HLA class I complexes (6,11–14). EGFR antagonism can also increase expression of HLA class I expression (14,15) and pro-inflammatory cytokines (16). We have recently shown that SHP2, which operates downstream of EGFR and dephosphorylates p-STAT1, plays an important role in HLA-induced immune escape in HNC (17). Thus, we evaluated if the EGFR-SHP2-STAT1 pathway might regulate HLA downregulation in HNC. The clinical significance of EGFR-induced HLA class I downregulation is important, since recently induction of anti-EGFR T cells has been demonstrated in cetuximab-treated HNC patients (18,19), supporting the crucial role for tumor cell downregulation of HLA antigen presentation by EGFR in evading CTL elimination. Thus, the goal of this study was to investigate the mechanism by which EGFR activation inhibits STAT1 activation as well as the HLA class I APM pathway and resulting adaptive antitumor immunity. We also exploited EGFR inhibition in cetuximab-treated HNC patients as a strategy by which this immune escape mechanism can be counteracted, linking HLA upregulation with clinical response in a novel phase II trial of neoadjuvant cetuximab therapy.
Materials and Methods
Cell lines
JHU-022, JHU-028, JHU-029 were a kind gift from Dr. James Rocco (Harvard Medical School, Boston, MA) in January 2007. SCC90, PCI-13, PCI-15B were isolated from patients treated at University of Pittsburgh Cancer Institute, Pittsburgh, through the explant/culture method, authenticated, and validated as unique using STR profiling and HLA genotyping every six month (20,21). 93-VU-147 T was a kind gift from Dr. Henning Bier (Technische Universitat Munchen, Munich, Germany) in October 2013. MCF-7 was a kind gift from Dr. Soldano Ferrone (Massachusetts General Hospital, Harvard Medical School, and Boston, MA) in December 2012. 2FTGH (STAT1+/+) and U3A (STAT1−/−) were a kind gift from Dr. George Stark (Cleveland Clinic Foundation, Cleveland, Ohio) in December 2011. All cell lines were routinely tested and found to be free of Mycoplasma. All cell lines were cultured in IMDM GlutaMAX media with antibiotics (Penicillin, 100 U/ml, Streptomycin 100 µg/ml) (Life Technologies, Grand Island, NY), 10% FBS (Mediatech, Herndon, VA).
Patients and specimens
All patients signed an informed consent approved by the Institutional Review Board (IRB #99-06). Peripheral venous blood samples were obtained from HNC patients with stage III/IVA disease (Table 1), receiving neoadjuvant cetuximab on a prospective phase II clinical trial (UPCI # 08-013, NCT 01218048). Tumors were biopsied immediately before, and again after 4 weeks of cetuximab therapy. Clinical response was analyzed by comparing paired CT scans pre/post cetuximab, and quantifying tumor measurement by a dedicated head and neck radiologist blinded to patient status. Anatomic tumor measurements were recorded in two dimensions and the cohort segregated into clinical “responders,” who demonstrated reduction in tumor volume, or “non-responders,” whose tumors grew during this therapy.
Table 1.
UPCI 08-013 Patient Demographics table 1
| No. of Patients | Tumor Site | Mean Age (yrs) | Males | Females | |
|---|---|---|---|---|---|
| 40 | OC | 19 | 58.6 | 30 | 10 |
| OP | 15 | ||||
| L | 6 | ||||
| HP | |||||
| Other Unknown primary |
|||||
Cytokines and antibodies
IFNγ, InterMune (Brisbane, CA) and rhEGF R&D Systems, (Minneapolis, MN) were purchased. LMP-2 (clone SY-1), TAP1 (NOB1), TAP2 (NOB2), Tapasin (clone TO-3), calreticulin (TO-11), HLA-A (clone LGIII-147.4.1) and HLA-B/C (clone B1.23.2) were characterized previously (22). FITC-conjugated HLA-A/B/C (clone G46-2.6 or clone W6-32), APC-conjugated β-2m (clone 2M2), PE-conjugated IFNγ receptor α chain (clone GIR-208), p-STAT1 staining used PE-conjugated anti-p-STAT1 (Tyr701), APC-conjugated anti-STAT1 Ab (BD Biosciences), anti-STAT1 (C-24) polyclonal (pAb) (Santa Cruz Biotech, Santa Cruz, CA), anti-β-actin mAb (Sigma-Aldrich Inc, St. Louis, MO).
Quantitative Real-Time PCR (qPCR)
RNA from HNC cells was extracted using Trizol (Invitrogen, Life Technologies, Carlsbad, CA), purified by RNA-cleanup (Qiagen, Gaithersburg, MD), Random hexamers, MuLV enzyme was used for cDNA synthesis (Applied Biosystems, Foster City, CA). PCR pre-developed probe for SHP2, HLA-B, STAT1, TAP-1 and β-actin were purchased from Applied Biosystem for TaqMan® Gene Expression Assay. Real-time PCR (7700 Real-Time PCR System; Applied Biosystems, Carlsbad, CA) used the following conditions: denaturation at 95°C for 10s, annealing at 60°C for 15s, and extension at 72°C for 30s. An initial denaturation step at 95°C for 5 min and final extension step at 72°C for 10 min were also included. Relative expression of the gene to endogenous control gene (β-actin/GUS) was calculated using the ΔCT method: relative expression = 2−ΔCT, where ΔCT = CT (shp2) − CT (β-actin/gus).
Immunoblotting
HNC cells were lysed in 1% Triton X-100 buffer with 1 mM PMSF, vortexed and centrifuged at 4°C, 16,100 g for 15 minutes. The supernatant protein was normalized and 40–60 µg of protein were size fractionated through a 4–12% SDS-PAGE gel (Lonza, Rockland, ME), transferred to a PVDF membrane (Millipore, Billerica, MA) and immunoblotted.
Immunohistochemistry (IHC)
Slides were deparaffinized and rehydrated using a standard histology protocol. Diva Retrieval solution (Biocare Medical, Concord, CA) and a Decloaking chamber at 124°C, 3:00 minutes, and cooled. The slides were placed on an Autostainer Plus (Dako, Carpenteria, CA) using 3% H2O2 for 5 minutes, CAS Block (Invitrogen, Grand Island, NY) for 10 minutes, the HLA-A (HCA2 mAb) and HLA-B/C (HC-10 mAb) Abs were applied using a 1:6000 dilution for 30 minutes. Both HLA-A and -B/C staining were quantified by positive pixel count v9 algorithm (Aperio). IHC analysis using a semiquantitative scale (H-score) comprising % tumor cells positive multiplied by intensity (0–3 scale) was performed without prior knowledge of clinicopathologic data (minimum score=0, maximum score=300).
Chromatin immunoprecipitation (ChIP) assay
Starved HNC cells (24 h, in AIM-V) prior to treatment (48h) were fixed with formaldehyde (1%, 10 min) (Sigma-Aldrich Inc.), quenched with glycine (0.125M) (Sigma-Aldrich Inc.), washed with PBS, and harvested. After centrifugation at 16,100g x12min, at 4°C, cells were lysed in SDS lysis buffer (Millipore) containing protease inhibitors. Chromatin was sheared by sonication for 12s at 20% of maximum (Cole Parmer Instrument) to fragment DNA. STAT1 and IgG control mAbs were used to immunoprecipitate STAT1-bound chromatin (10µg of Ab), at 4°C. Protein-DNA crosslinks were reversed at 65°C overnight. Initial denaturation: 95°C for 3 minutes, denaturation: 95°C for 15s, anneal and extension: primer-specific temp for 60s; repeat steps b and c for 40 cycles. Commercially available “SimpleChIP® Human TAP1 Promoter primer” (Cell signaling, Inc.) was used to amplify canonical sequence in the TAP1 promoter for STAT1 binding.
51Cr cytotoxicity assay
Cytotoxicity was determined using a 4h 51Cr release assay. Untreated and IFNγ-treated JHU-029 HNC cells were incubated in 100 µL of RPMI 1640 media with 25 µCi of Na251CrO4 (Perkin Elmer, Boston MA) for 1h at 37°C and resuspended in RPMI 1640 media. Cells were washed (2X) and plated at indicated effector/target ratio (E/T ratio 40:1) in U-bottom 96-well plates. Cetuximab (10 µg/ml), anti-HLA class I mAb (50 µg/ml) was added in NK cell: JHU-029 coculture. Plates were incubated for 4h at 37°C in a 5% CO2 atmosphere. Controls for spontaneous (cells only) and maximal lysis (cells treated with 5% Triton-X) were included. Each reaction was performed in triplicate. 50 µl supernatants were collected and analyzed with a Perkin Elmer 96-well plate gamma counter. Results were normalized with the formula of specific lysis = (experimental lysis − spontaneous lysis)/(maximum lysis − spontaneous lysis) × 100.
siRNA transfections
HNC transfected with SHP2 or non-targeting siRNA control (Ambion, Austin, Tx) and lipofectamine-RNAi max (Life Technologies), and Opti-MEM I (Life Technologies) according to the Lipofectamine-RNAi max instructions. SiRNA used in this assay. SHP2 SiRNA: 5’-GGAGAACGGUUUGAUUCUUTT-3’ (s) and 5’-AAGAAUCAAACCGUUCUCCTC-3’ (as) EGFR SiRNA: Custom siRNA oligo for wt EGFR, AACUCUGGAGGAAAAGAAAGUdTdT Control SiRNA: 5’-AGUACAGCAAACGAUACGGtt-3’ (s) and 5’-CCGUAUCGUUUGCUGUACUtt-3’.
Statistical Analysis
Data were analyzed statistically using GraphPaD Prism 4.0. A two-tailed unpaired or paired t-test was used to calculate whether observed differences were statistically significant, defined as p<0.05.
Results
Cetuximab-mediated EGFR inhibition differentially enhances expression of HLA class I alleles and APM components in a STAT1-dependent fashion
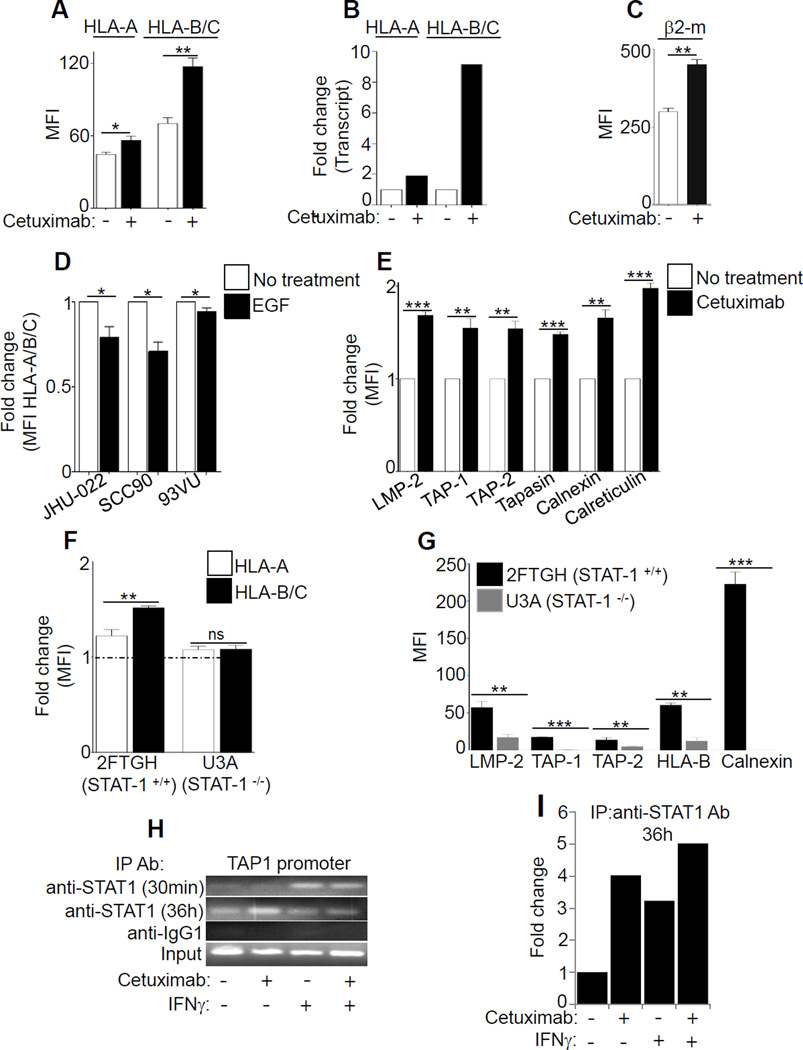
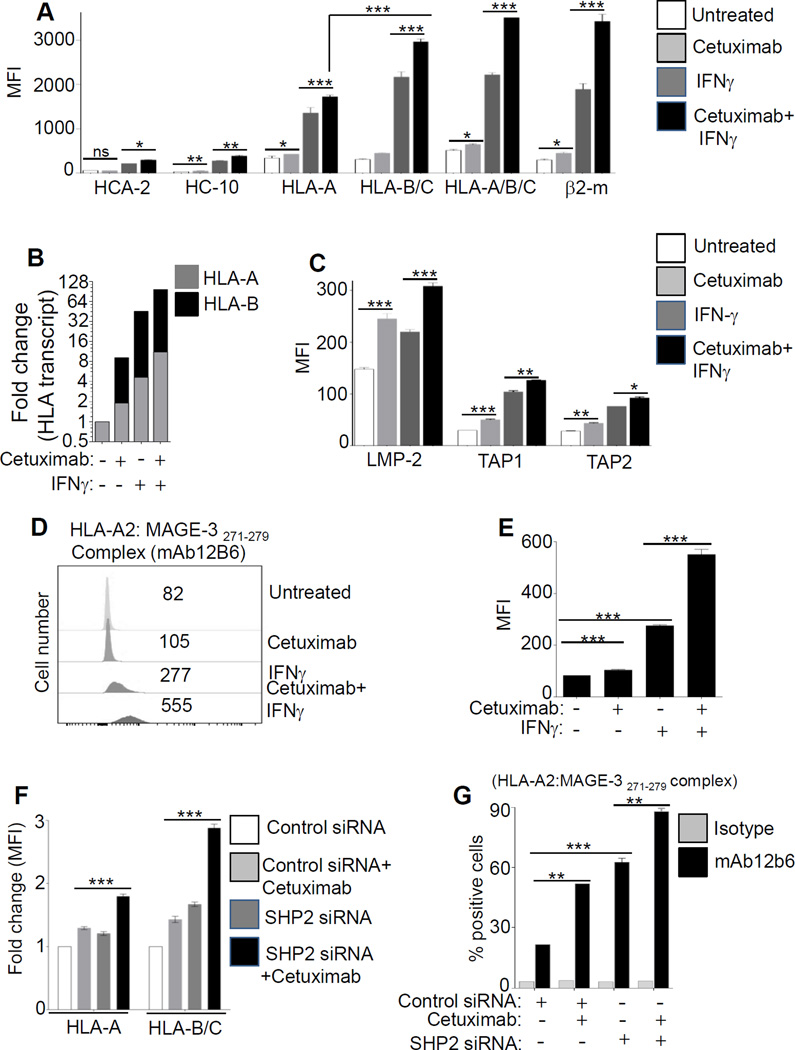
First, we determined the effect of EGFR signaling on the level of expression of two distinct alleles, HLA-A or HLA-B. Interestingly, incubation of HNC cells with the EGFR inhibitor cetuximab (10 µg/ml, 48h) led to increased HLA-B expression (~2 fold for protein, 9 fold increased transcription), to a greater extent than the expression of HLA-A (1.25-fold protein level, 2 fold increase in transcription) (Fig. 1A-B). Similar upregulation was observed with β2-m expression after EGFR inhibition (Fig. 1C). Although EGFR inhibition did not enhance free surface HLA-A heavy chains (HCA-2 mAb), elevated levels of free HLA-B/C heavy chains (HC-10 mAb) was observed (Supplementary Fig. S1A). Similarly, EGF stimulation inhibited HLA class I expression, inducing inhibition of HLA-B to a greater extent than of HLA-A (Fig. 1D, Supplementary Fig. S1B). In addition to upregulating HLA class I molecules, cetuximab treatment enhanced levels of intracellular APM components LMP2, TAP1/2, tapasin, calnexin and calreticulin (Fig. 1E).
Figure 1. Cetuximab-mediated EGFR inhibition differentially enhances expression of HLA class I alleles and APM components in a STAT1-dependent fashion.
JHU-029 HNC cells were left untreated or were treated for 48h with the EGFR inhibitor mAb cetuximab (10 µg/ml). Levels of HLA-A alleles or HLA-B/C alleles were determined by FACS (A) or by qPCR (B), and levels of surface β2-m were measured by FACS (C). HNC cells were left untreated or were treated for 48h with the rhEGF (10 ng/ml), and levels of HLA class I (mAb W6/32) were determined by FACS (D). The levels of intracellular LMP2, TAP1, TAP2, tapasin, calnexin, and calreticulin (E) were measured by FACS. In Figure 1F, levels of HLA-A and HLA-B/C were evaluated in parental 2FTGH (STAT1+/+) and derivative U3A (STAT1−/−) cells after treatment with EGFR siRNA plus cetuximab or control siRNA. In Figure 1G, levels of intracellular LMP2, TAP1, TAP2, surface HLA-B/C, calnexin were examined in 2FTGH (STAT1+/+) and U3A (STAT1−/−) cells. MFI values (EGFR siRNA-control siRNA) were determined by FACS. In Figure 1H-I, cetuximab-induced STAT1 binding to the GAS element (gamma interferon activation site) of the TAP1 promoter was measured using a chromatin immunoprecipitation (ChIP) assay. JHU-029 cells were treated with cetuximab (10 µg/ml for 30 min or 36 h), IFNγ (10 U/ml) and cetuximab plus IFNγ (10 µg/ml, 10 U/ml) and enhanced binding of STAT1 at TAP1 promoter was determined by chip assay. Results represent mean +/− SEM from three independent experiments. (*P≤0.05, **P≤0.001, ***P≤0.0001)
We next found that EGFR inhibition led to preferential upregulation of HLA-B/C vs HLA-A molecules, but only in STAT1+/+ cells (Fig. 1F). Similar results were observed for expression APM components LMP2, TAP1/2, and calnexin, indicating the STAT1 dependence of EGFR-driven HLA/APM downregulation (Fig. 1G). Furthermore, EGFR siRNA increased STAT1 activation in STAT1+/+ cells, but not in STAT1−/− cells (Supplementary Fig. S1C-D). Consistent with these findings, the STAT1 inhibitor fludarabine (20µM) (23,24) abrogated the effect of treatment of HNC cells with cetuximab, IFNγ, or cetuximab plus IFNγ, while not affecting EGFR expression (Supplementary Fig. S1E). A ChIP assay confirmed that cetuximab indeed induced binding of STAT1 to the TAP1 promoter (GAS element), providing physical evidence for transcriptional activation of APM pathway genes (Fig. 1H-I, Supplementary Fig. S1F).
Inhibition of EGFR-SHP2 pathway induces STAT1 activation
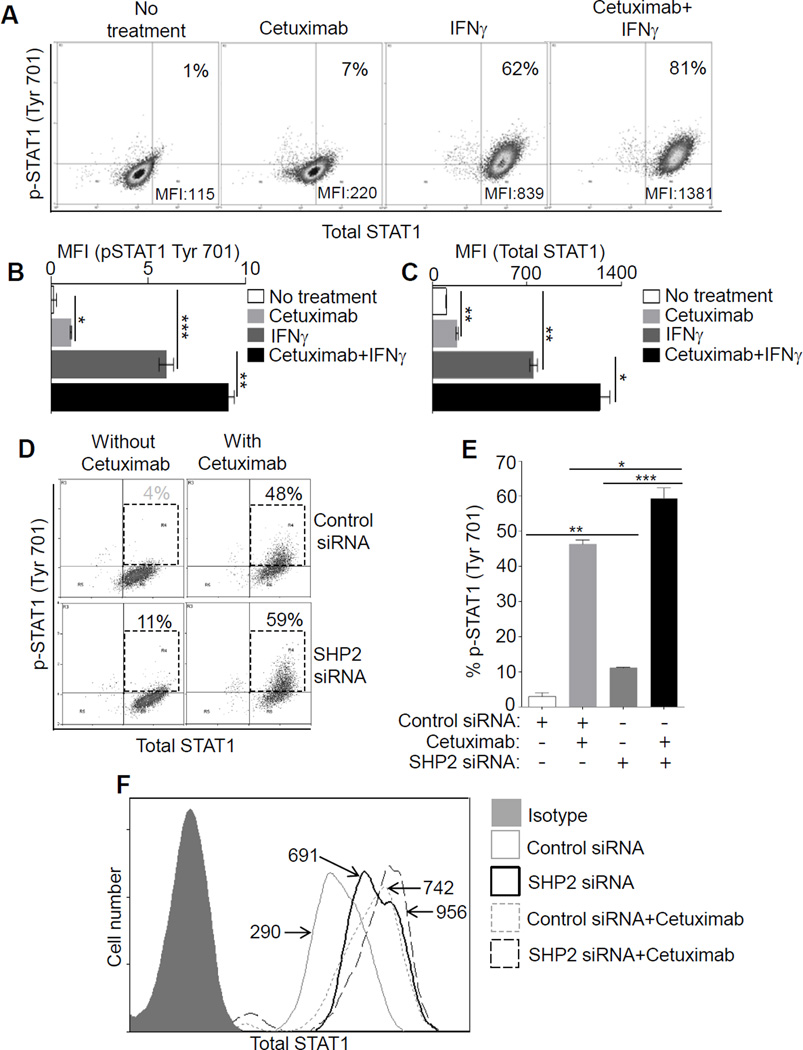
Having shown that EGFR can regulate HLA expression in a STAT1-dependent fashion, we hypothesized that HLA downregulation due to autocrine or paracrine EGFR activation may drive SHP2 phosphatase activation and resulting STAT1 suppression (13,17) . To investigate this possibility, we treated JHU-029 HNC cells with cetuximab (10 µg/ml, 24 hours) and found that cetuximab treatment significantly decreased SHP2 expression, whereas IFNγ treatment (10 IU/ml) as a positive control had no effect on SHP2 expression. The combination of cetuximab and IFNγ reduced SHP2 levels, when compared with untreated HNC cells (Supplementary Fig. S2A). We then measured the expression of p-STAT1 (Tyr701) and total STAT1 after cetuximab alone or plus IFNγ treatment. After cetuximab treatment a slight increase in the level of p-STAT1 (Tyr701) was observed (from 1% to 7% p-STAT1+ cells), while a more prominent increase in the level of total STAT1 was observed (~2 fold higher MFI vs. untreated). IFNγ treatment strongly increased expression of both p-STAT1 (Tyr701) (1% to 62%) and total STAT1 (~7.9 fold increase in MFI vs. untreated). Interestingly, cetuximab treatment augmented the ability of IFNγ to induce p-STAT1 (from 62% to 81% positive cells) and total STAT1 (~12 fold higher MFI vs. untreated) (Fig. 2A-C). A similar observation was confirmed with immunoblotting (Supplementary Fig. S2B), and STAT1 transcript analysis (Supplementary Fig. S2C). We also evaluated STAT1 and HLA expression after co-inhibiting SHP2 and EGFR (using siRNA or cetuximab) in order to bypass EGFR. SHP2 phosphatase depletion using siRNA (17) enhanced cetuximab-induced p-STAT1 (Tyr701), as well as total STAT1, expression suggesting that high SHP2 expression downstream of EGFR in HNC cells prevents STAT1-mediated signaling. Indeed, the combination of SHP2 siRNA and cetuximab treatment strongly enhanced cetuximab-mediated STAT1 upregulation (Fig. 2D-F, Supplementary Fig. S2D).
Figure 2. Inhibition of EGFR-SHP2 pathway induces STAT1 activation.
In JHU-029 cells, levels of p-STAT1 (Tyr701), total STAT1 were determined after treatment (48h) with cetuximab (10 µg/ml), IFNγ (10 U/ml), cetuximab plus IFNγ (10 µg/ml, 10U/ml), and a representative dot plot analysis with upper right quadrants indicating % p-STAT1 (Tyr701) cells (y-axis) and MFI values of total STAT1 (x-axis) is shown (A). Statistical analysis for p-STAT1 (Tyr 701) MFI and total STAT-1 MFI are shown (B-C). HNC cells were treated with SHP2 siRNA or control siRNA (24h). Afterward cells were treated with cetuximab (5 µg/ml, 48h) and levels of p-STAT1 (% positive cells, y-axis) and total STAT1 (MFI, x-axis) were determined and a dot plot analysis is shown (D). Numbers in UR quadrants of dot plot analysis for p-STAT1 (Tyr701) is also shown in a separate bar diagram (E). Similarly, levels of total STAT1 in JHU-029 cells were determined with FACS in the experimental conditions indicated in the histogram: isotype Ab (filled gray), control siRNA (gray thin solid line, MFI 290), SHP-2 siRNA (black thick solid line, MFI 691), control siRNA+ cetuximab (gray dotted line, MFI 742), SHP-2 siRNA+ cetuximab (black thin, long dashed line, MFI 956) (F). Results represent mean +/− SEM from three independent experiments. (*P≤0.05, **P≤0.001, ***P≤0.0001)
Cetuximab treatment increases IFNγ receptor 1 expression
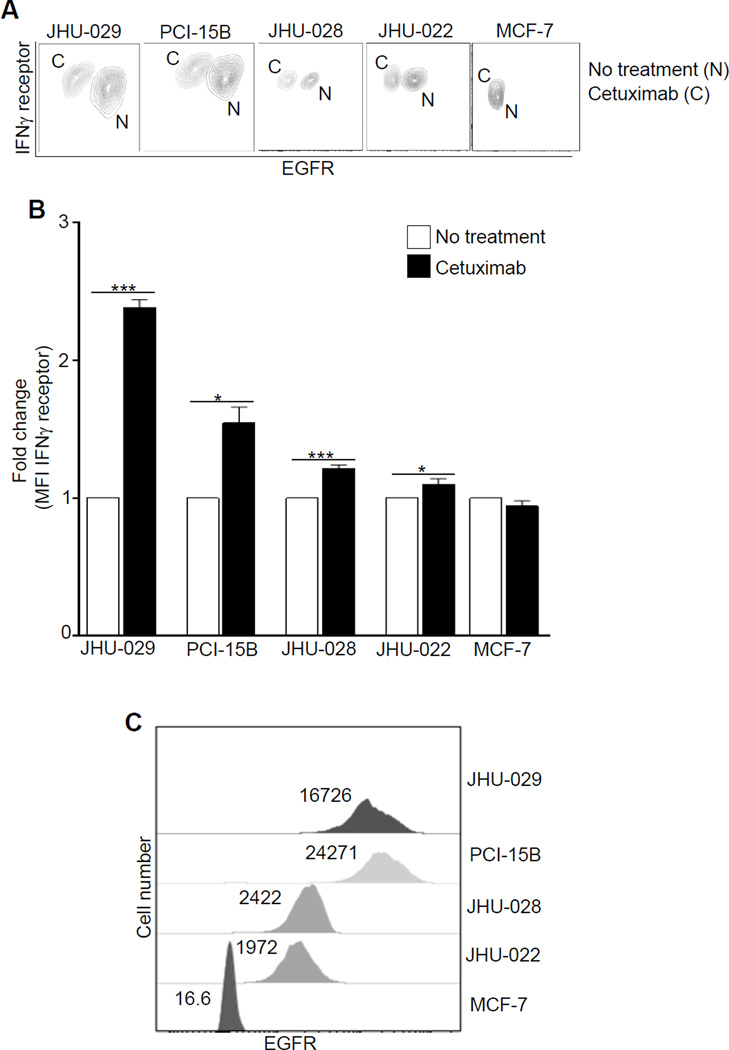
Since EGFR signaling might downregulate IFNγ receptor1 (IFNRI) (25), providing a mechanism for synergistic effects of EGFR blockade with IFNγ for STAT1-mediated HLA upregulation, we evaluated levels of IFNRI after cetuximab treatment. Indeed, cetuximab increased the expression of IFNRI in several HNC cell lines tested, in an EGFR-dependent fashion (Fig. 3A-C). To test the hypothesis that EGFR density is important in regulating IFNRI, we used cetuximab and EGFR siRNA to abolish EGFR-proximal signaling (Supplementary Fig. S2E), which independently showed an increase of IFNRI expression (Supplementary Fig. S2F). As expected, the combination of cetuximab treatment plus EGFR siRNA knockdown showed most pronounced effect on IFNRI downregulation (Supplementary Fig. S2F).
Figure 3. Cetuximab treatment increases IFNγ receptor 1 expression.
JHU-029, PCI-15B, JHU-028, JHU-022, EGFR+ HNC cells, and MCF-7 EGFRlow/− breast cancer cells were incubated with cetuximab (10 µg/ml, 72h) and levels of IFNγ receptor α chain (CD119) (A-B), and EGFR expression (A,C) were determined by FACS. Results represent mean +/− SEM from three independent experiments. (*P≤0.05, ***P≤0.0001)
Cetuximab-activated NK cells and IFNγ increase expression of HLA class I APM pathway
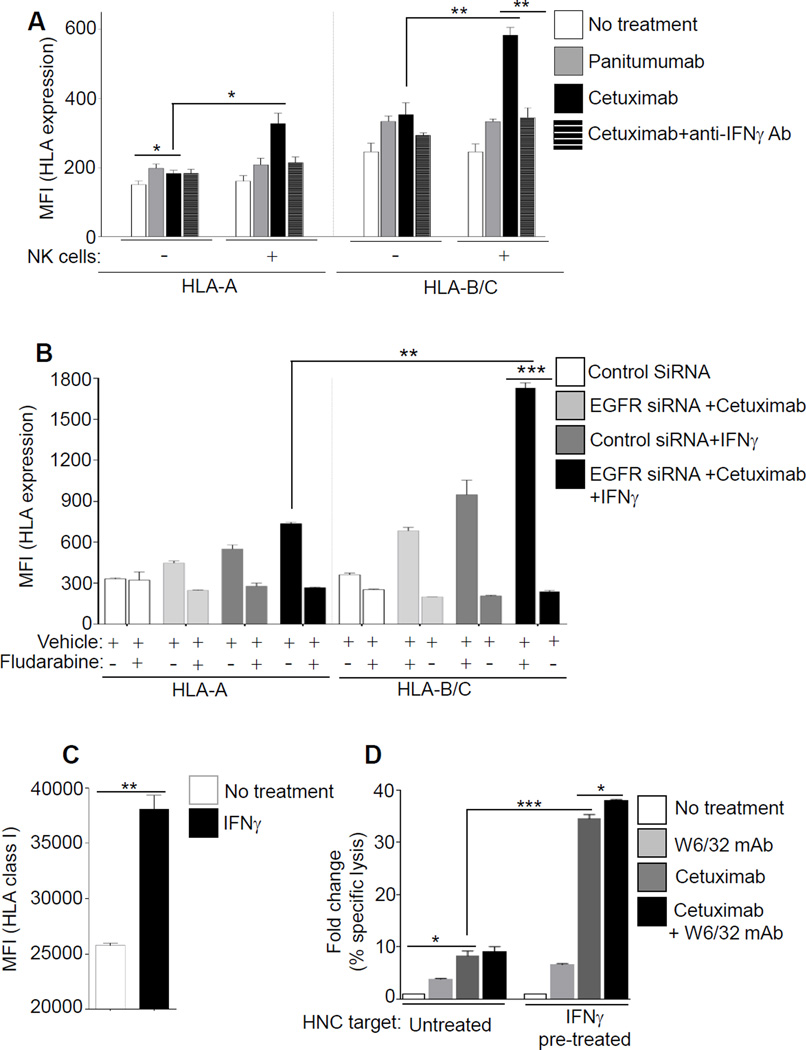
Because IFNγ increases expression of HLA class I and APM components in HNC cells, and since NK cells secrete IFNγ after recognizing cetuximab-coated HNC cells in the tumor microenvironment (18,26), we considered the impact of cetuximab-activated NK cells during co-culture with HNC cells (19,27). Under these conditions, even more robust expression of HLA-A and HLA-B/C was observed (Fig. 4A), particularly when both NK cells and cetuximab were present. As shown, IFNγ released from cetuximab-activated NK cells further evoked HLA-A and HLA-B/C upregulation, since an IFNγ neutralizing Ab abrogated the beneficial effect of NK cell treatment in both cases (Fig. 4A). An IgG2, anti-EGFR mAb panitumumab failed to activate NK cells under similar conditions (19). Again, HLA-B/C alleles showed a more pronounced enhancement after cetuximab treatment or by cetuximab-activated NK cells (~1.93-fold and ~ 1.78-fold induction), in comparison to HLA-A. In support of a common pathway, the STAT1 inhibitor fludarabine (23) abrogated HLA-A and HLA-B/C upregulation in response to cetuximab, EGFR siRNA, or IFNγ treatment (Fig. 4B). We further evaluated the contribution of IFNγ-induced HLA class I expression to NK cell-mediated antitumor effects (Fig. 4C). Cetuximab-mediated antibody-dependent cellular cytotoxicity (ADCC) was significantly enhanced against IFNγ–treated HNC targets, and blocking HLA class I with W6/32 mAb (pan-HLA class I mAb) augmented cetuximab-mediated ADCC (Fig. 4D).
Figure 4. Cetuximab-activated NK cells and IFNγ increase expression of HLA class I APM pathway.
(A) JHU-029 HNC cells were cultured alone, or JHU-029 plus NK cells in co-culture (1:1 ratio) were left untreated or were treated for 48h with panitumumab (IgG2, 10 µg/ml), cetuximab (IgG1, 10 µg/ml). Levels of HLA-A (left panel) or HLA-B/C (right panel) were determined by FACS. In parallel, polyclonal anti-IFNγ Ab (10 µg/ml) was added at indicated conditions to determine the effect of IFNγ released from cetuximab-activated NK cells. (B) HNC cells were pre-treated with fludarabine (20 µM), and after EGFR siRNA or cetuximab treatment (48h, 10 µg/ml) levels of surface HLA-A, HLA-B/C were determined by FACS. (C) JHU-029 HNC cells were cultured alone, or were treated with IFNγ (10 U/ml, 36h) and enhanced levels of HLA class I (mAb W6/32) was verified with FCAS. (D) NK cell cytotoxicity (4h 51Cr release assay, 40:1 E/T ratio) against untreated or IFNγ pre-treated HNC targets (C), were independently evaluated in presence of mAb W6/32 (50 µg/ml), cetuximab (10 µg/ml) or combination of mAb W6/32 plus cetuximab. The ratio of NK cell cytotoxicity against untreated HNC targets, and IFNγ-treated HNC targets is shown. Results represent mean +/− SEM from three independent experiments. (*P≤0.05, **P≤0.001, ***P≤0.0001)
SHP2 inhibition robustly enhances cetuximab-mediated tumor-antigen (TA) presentation
Next, we evaluated the combined effect of cetuximab and IFNγ on the expression of free HLA-A (HCA-2 mAb) or free HLA-B (HC-10 mAb), surface HLA-A and HLA-B/C, surface pan-HLA class I (HLA-A/B/C), or β2-m. Cetuximab alone increased ~1.25-fold expression of HLA-A in comparison to ~1.45-fold increase for HLA-B/C (Fig. 5A), IFNγ alone increased ~3.9-fold expression of HLA-A in comparison to a ~6.9-fold increase for HLA-B/C when compared to that of no treatment. The most prominent upregulation of HLA-A and HLA-B/C (~ 5 fold and ~9.5 fold vs. untreated, p<0.0001) was observed when the combination of IFNγ and cetuximab was used. We also found greater HLA-B versus HLA-A allele transcripts after treatment with cetuximab, with IFNγ alone, or with cetuximab plus IFNγ (Fig. 5B, and Supplementary Fig. S3A-E).
Figure 5. SHP2 inhibition robustly enhances cetuximab-mediated TA presentation.
JHU-029 cells were treated with cetuximab (10 µg/ml, 72h), IFNγ (10U/ml, 72h), cetuximab plus IFNγ (10 µg/ml, 10U/ml respectively, 72h) and levels of intracellular free HLA class I A or B, HLA-A, HLA-B/C, HLA-A/B/C, β2-m were determined by FACS (A). Under above described conditions the transcript levels of HLA-A and HLA-B were determined by qPCR (B). Similarly, in the above described conditions, levels of LMP2, TAP1, and TAP2 were determined by FACS (C). In HLA-A2+ JHU-022 cells presentation of HLA-A2:MAGE-3271–279–peptide-complex (probed by mAb 12B6, followed by FITC-(Fab)2 was determined with flow cytometry after treatment with cetuximab (10 µg/ml), IFNγ (10 U/ml), cetuximab plus IFNγ (10 µg/ml, 10 U/ml respectively, 72h). A representative histogram showing MFI values (D) and graphical presentation (E) is shown. HNC cells JHU-029 (F) were treated with SHP2 siRNA or control siRNA. After 24h cells were treated with cetuximab (10 µg/ml, 48h) and levels of HLA-A and HLA-B/C were evaluated by FACS. Levels of HLA-A2:MAGE-3271–279–peptide-complex (clone mAb 12b6) presentation were determined by FACS after treatment with control siRNA, control siRNA plus cetuximab (5µg/ml), SHP2 siRNA and SHP2 siRNA plus cetuximab (additional 48h) in MAGE-3271–279 TA positive HLA-A2+- PCI-13 (G). Results represent mean +/− SEM from three independent experiments. (*P≤0.05, **P≤0.001, ***P≤0.0001)
Because cetuximab treatment enhanced the expression of LMP2, TAP1/2 (Fig. 5C, Supplementary Fig. S4A-B), we investigated whether the enhanced HLA class I APM components enhanced surface presentation of tumor antigens (TA). We utilized a novel mAb (12b6), recognizing the HLA-A2:MAGE-3271–279 complex, (Supplementary Fig. S5A-B), to quantitatively measure levels of surface HLA-TA complexes. Cetuximab enhanced HLA-A2:MAGE-3271–279 complex (p<0.001) (Fig. 5D-E), which was even more robust after IFNγ treatment (p<0.0001). Interestingly, the combination of cetuximab and IFNγ treatment evoked the highest level of HLA-A2:MAGE-3271–279–peptide complexes (p<0.0001). Indeed, the combination of SHP2 siRNA and cetuximab treatment strongly enhanced cetuximab induced HLA-A and HLA-B/C expression, most prominently in the latter alleles (Fig. 5F, Supplementary Fig. S5C). SHP2 depletion in HLA-A2+ HNC cells also enhanced HLA-A2:MAGE-3271–279-peptide presentation after cetuximab treatment (Fig. 5G, Supplementary Fig. S5D), whereas no binding was observed in HLA-A2− or TA− HNC cells (Supplementary Fig. S5E-F).
Cetuximab neoadjuvant therapy enhances expression of HLA class I in HNC patients
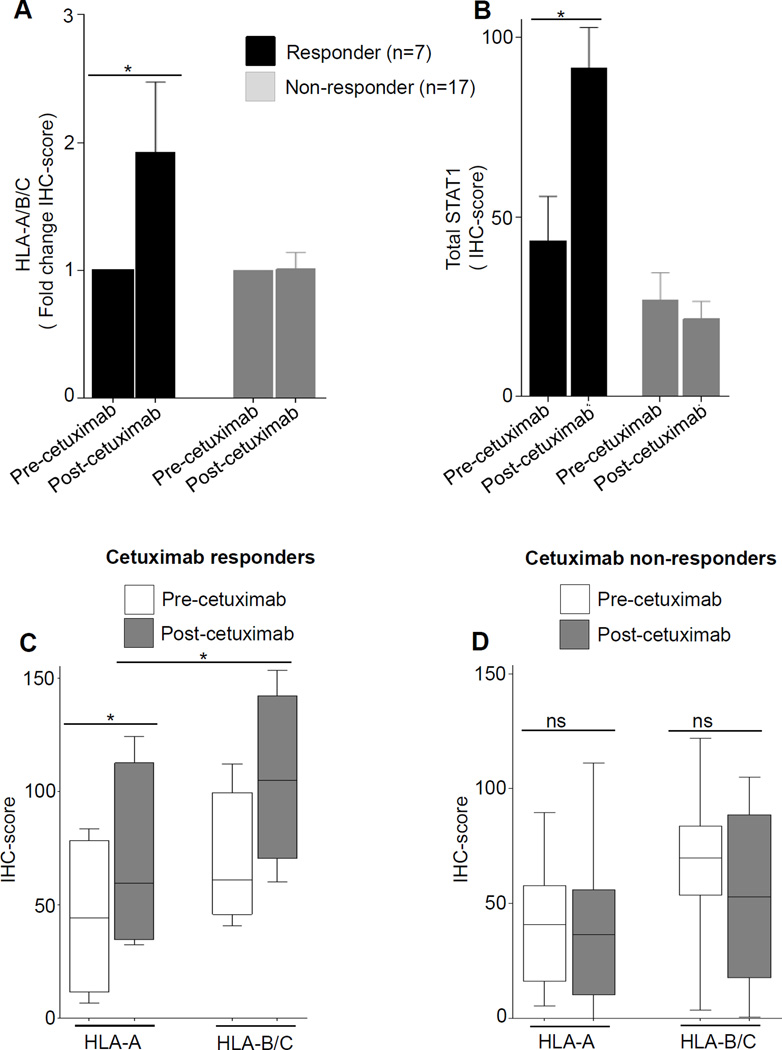
In a novel phase II prospective clinical trial, tumors from HNC patients were biopsied before and after 4 weeks of single-agent neoadjuvant cetuximab therapy. HLA class I expression was measured semi-quantitatively using IHC and digital image analysis, and correlated with clinical response by paired pre/post CT scans to identify clinical “responders.” After cetuximab therapy, both HLA alleles, and STAT-1 were upregulated in the clinical responders (n=7) but not in non-responders (n=17) (Fig. 6 A-D) to EGFR-specific mAb therapy.
Figure 6. Cetuximab neoadjuvant therapy enhances HLA class I expression in patients’ tumors in clinical responder but not in nonresponder HNC patients.
Tumor specimens from HNC patients (n=24) enrolled on UPCI trial 08-013 were biopsied pre-treatment and post-neoadjuvant treatment (cetuximab IV 400mg/m2 day 1 then 250 mg/m2 alone days 8, 15, and 22), and a tissue microarray was prepared from paired tumor biopsies. Expression of HLA-A (mAb HCA2), HLA-B (mAb HC10), and STAT1 (mAb C-24) were determined in neoadjuvant cetuximab-treated HNC patients by semiquantitative IHC (A-D), and correlated with tumor shrinkage (CT scan “responders”) after 4 weeks of cetuximab therapy. (* P≤0.05)
Discussion
In HNC, low levels of HLA class I and APM component expression preclude effectiveness of CTL responses in mediating tumor elimination (11), and this mechanism of immune escape is a consequence of diminished STAT1 activation generated by the overexpression of SHP2 (17). Multiple pathways are linked with SHP2 functions in HNC, primarily the EGFR-SHP2 pathway. Due to the frequent overexpression of EGFR, which is a poor prognostic factor in HNC, constitutive activation of this pathway may greatly facilitate and ‘immune-escape’ phenotype through suppression of p-STAT1-mediated expression of the HLA/APM pathway. This study sheds light on the mechanism(s) responsible for the diminished TA processing and presentation due to suppression of STAT1 and HLA class I APM components in HNC, which may be reversed through EGFR blockade, IFNγ release due to cetuximab-activated NK cells, or both. The effect is likely to have a beneficial impact on the clinical course of the disease in HNC patients treated with cetuximab.
Recently, we have shown that in HNC patients, cetuximab induces cross-priming of EGFR-specific CTLs by NK:DC cross-talk (18,19). However, the determinants of TA recognition by CTLs may benefit cetuximab-mediated clinical responses. Intriguingly, processing and presentation of HLA class I peptide-complex is an intricate-process (28), and polymorphism in HLA class I alleles (29), their differential levels and dynamic role of APM components represent important immune escape mechanisms from adaptive immunity in cancer (19,29). Using a novel HLA-A2:MAGE-3-specific mAb, we demonstrated quantitatively enhanced TA presentation in HNC cells, which is critical for CTL lysis. Thus, the likelihood of generation of greater repertoire of TAs (“antigen spreading”) appears to result from EGFR blockade using cetuximab, perhaps due to IFNγ-induced antigen presentation plus upregulated HLA alleles. Enhanced recognition of the peptide-HLA-A2 complex using the combination of cetuximab and IFNγ could be monitored diagnostically as a measure of Th1-biased immune responses. However, in light of superior restoration of HLA-B with cetuximab, greater characterization of HLA-B-restricted TAs is also warranted, particularly during cetuximab-based immunotherapy. These effects could be overcome by a greater drop in HLA class I expression after EGF treatment of HNC cells. The effect of HLA upregulation on reduced cetuximab-mediated ADCC supports a moderate impact of NK cell inhibitory, killer immunoglobulin-like receptors (KIR) and the tumor cell/HLA class I interaction during cetuximab-mediated ADCC.
Polymorphism of HLA class I alleles may play a dominant role in regulating NK-cell effector function. Interaction of few NK-cell inhibitory receptors with specific HLA allele plays important role in NK cell-mediated antitumor responses. Inhibitory KIRs have higher affinity for HLA-B than for HLA-A (29). This suggests the strong possibility that HLA-B, which interacts with KIR, may have negative impact of NK-cell stimulation (29), and HNC may become more resilient to further NK-cell attack (30,31). HLA-B is most often loaded with antigenic peptides (32). In HNC cell lines used in our study, IFNγ induces HLA-B/C more strongly than HLA-A (32,33), rarely reported previously. This observation reflects the fact that EGFR signaling in combination with p-STAT1 suppression by SHP2 have overwhelmingly negative effects on immunogenic TA-peptide presentation, abrogating the generation, loading and presentation on surface HLA class I:TA peptide complexes, which are necessary for CTL lysis.
Cetuximab induces EGFR-specific CTL responses in some HNC patients (18), whereas clinical response to cetuximab is only observed in a subset of patients (~20%) (34,35). The effects of concerted antitumor immune responses involving NK cells, NK-DC cross-talk and CTL responses (18,19,36,37), along with upregulation of HLA class I and IFNRI may contribute to cetuximab therapy. Indeed, we observed STAT-1, HLA class I upregulation in cetuximab-treated patients in a novel neoadjuvant trial, suggesting that EGFR inhibition and/or IFNγ release contributes to the reversal of HLA downregulation. Results from recent studies also indicated an immunosuppressive effect of EGFR signaling on STAT1-dependent HLA class I and CIITA genes (38,39). However, HLA class I and CIITA are modulated by both total STAT1 and p-STAT1 (40,41), and a recent report also indicates that EGFR inhibitors reduces PD-L1 expression in lung tumors (42), suggesting multiple immune escape mechanisms mediated by EGFR signaling.
Supplementary Material
Acknowledgments
We acknowledge patients and their families for participating in this study. We acknowledge excellent technical assistance from Clayton Mathis, Michael Meyer, and Bratislav Janjic, University of Pittsburgh Cancer Institute. We thank Ferris lab members for helpful suggestions.
Grant Support
This work was supported by National Institute of Health grants R01 DE19727, P50 CA097190, CA110249 and University of Pittsburgh Cancer Center Support Grant P30CA047904.
Abbreviations used in this article
- HNC
head and neck cancer
- EGFR
Epidermal growth factor receptor
- SHP2
Src-homology phosphatase2
- TA
tumor antigen
- STAT1
Signal Transducer and Activator of Transcription 1
- IFNAR
IFNγ receptor 1.
Footnotes
Disclosure of Potential Conflicts of Interest
RLF reports receiving commercial research grants from Bristol-Myers Squibb, Merck, and Astra Zeneca. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: R.M. Srivastava, S. Ferrone, RL Ferris
Acquisition of data: R.M. Srivastava, S. Trivedi, R. R. Seethala, H-B Jie, B F Branstetter, R L Ferris
Analysis and or interpretation of data: R.M. Srivastava, Trivedi S, F. Concha-Benavente, L Wang, H-B Jie, R. R. Seethala, S Ferrone, R.L. Ferris
Development of methodology: F. Concha-Benavente, R. R. Seethala, R L Ferris
Writing, review, and/or revision of manuscript: R.M. Srivastava, S. Ferrone, B F Branstetter, R.L. Ferris
Administrative, technical, or material support: R. R. Seethala, B F Branstetter,
Study supervision: R.L. Ferris
References
- 1.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. The New England journal of medicine. 2006;354(6):567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 2.Concha-Benavente F, Srivastava RM, Ferrone S, Ferris RL. EGFR-mediated tumor immunoescape: The imbalance between phosphorylated STAT1 and phosphorylated STAT3. Oncoimmunology. 2013;2(12):e27215. doi: 10.4161/onci.27215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollack BP. EGFR inhibitors, MHC expression and immune responses : Can EGFR inhibitors be used as immune response modifiers? Oncoimmunology. 2012;1(1):71–74. doi: 10.4161/onci.1.1.18073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. Journal of the National Cancer Institute. 2013;105(16):1172–1187. doi: 10.1093/jnci/djt184. [DOI] [PubMed] [Google Scholar]
- 5.Ferris RL, Hunt JL, Ferrone S. Human leukocyte antigen (HLA) class I defects in head and neck cancer: molecular mechanisms and clinical significance. Immunologic research. 2005;33(2):113–133. doi: 10.1385/IR:33:2:113. [DOI] [PubMed] [Google Scholar]
- 6.Ferris RL, Whiteside TL, Ferrone S. Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(13):3890–3895. doi: 10.1158/1078-0432.CCR-05-2750. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann F, Lehr HA, Drexler I, Sutter G, Hengstler J, Wollscheid U, et al. HER-2/neu-mediated regulation of components of the MHC class I antigen-processing pathway. Cancer research. 2004;64(1):215–220. doi: 10.1158/0008-5472.can-2522-2. [DOI] [PubMed] [Google Scholar]
- 8.Nouri A, Cannell H, Onguti M, Tezabwala B, Oliver R. The possible relevance of the expression of MHC antigens and of EGF receptor in aggressive oral tumours. International journal of oncology. 1997;10(6):1217–1222. doi: 10.3892/ijo.10.6.1217. [DOI] [PubMed] [Google Scholar]
- 9.Seliger B. Molecular mechanisms of MHC class I abnormalities and APM components in human tumors. Cancer immunology, immunotherapy : CII. 2008;57(11):1719–1726. doi: 10.1007/s00262-008-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimura K, Shiraishi K, Mueller A, Izawa S, Kua LF, So J, et al. The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J Immunol. 2013;191(12):6261–6272. doi: 10.4049/jimmunol.1301597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Albaitero A, Nayak JV, Ogino T, Machandia A, Gooding W, DeLeo AB, et al. Role of antigen-processing machinery in the in vitro resistance of squamous cell carcinoma of the head and neck cells to recognition by CTL. J Immunol. 2006;176(6):3402–3409. doi: 10.4049/jimmunol.176.6.3402. [DOI] [PubMed] [Google Scholar]
- 12.Trivedi S, Concha-Benavente F, Srivastava RM, Jie HB, Gibson SP, Schmitt NC, et al. Immune biomarkers of anti-EGFR monoclonal antibody therapy. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014 doi: 10.1093/annonc/mdu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer immunology, immunotherapy : CII. 2011;60(4):525–535. doi: 10.1007/s00262-010-0961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollack BP, Sapkota B, Cartee TV. Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(13):4400–4413. doi: 10.1158/1078-0432.CCR-10-3283. [DOI] [PubMed] [Google Scholar]
- 15.Lupberger J, Duong FH, Fofana I, Zona L, Xiao F, Thumann C, et al. Epidermal growth factor receptor signaling impairs the antiviral activity of interferon-alpha. Hepatology. 2013;58(4):1225–1235. doi: 10.1002/hep.26404. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher EV, Love-Homan L, Sobhakumari A, Feddersen CR, Koch AT, Goel A, et al. EGFR inhibition induces proinflammatory cytokines via NOX4 in HNSCC. Molecular cancer research : MCR. 2013;11(12):1574–1584. doi: 10.1158/1541-7786.MCR-13-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leibowitz MS, Srivastava RM, Andrade Filho PA, Egloff AM, Wang L, Seethala RR, et al. SHP2 is overexpressed and inhibits pSTAT1-mediated APM component expression, T-cell attracting chemokine secretion, and CTL recognition in head and neck cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(4):798–808. doi: 10.1158/1078-0432.CCR-12-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava RM, Lee SC, Andrade Filho PA, Lord CA, Jie HB, Davidson HC, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(7):1858–1872. doi: 10.1158/1078-0432.CCR-12-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SC, Srivastava RM, Lopez-Albaitero A, Ferrone S, Ferris RL. Natural killer (NK): dendritic cell (DC) cross talk induced by therapeutic monoclonal antibody triggers tumor antigen-specific T cell immunity. Immunologic research. 50(2–3):248–254. doi: 10.1007/s12026-011-8231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(23):7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer research. 1989;49(18):5167–5175. [PubMed] [Google Scholar]
- 22.Bandoh N, Ogino T, Cho HS, Hur SY, Shen J, Wang X, et al. Development and characterization of human constitutive proteasome and immunoproteasome subunit-specific monoclonal antibodies. Tissue antigens. 2005;66(3):185–194. doi: 10.1111/j.1399-0039.2005.00462.x. [DOI] [PubMed] [Google Scholar]
- 23.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nature medicine. 1999;5(4):444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 24.Fryknas M, Dhar S, Oberg F, Rickardson L, Rydaker M, Goransson H, et al. STAT1 signaling is associated with acquired crossresistance to doxorubicin and radiation in myeloma cell lines. International journal of cancer Journal international du cancer. 2007;120(1):189–195. doi: 10.1002/ijc.22291. [DOI] [PubMed] [Google Scholar]
- 25.Mitra RS, Nickoloff BJ. Epidermal growth factor and transforming growth factor-alpha decrease gamma interferon receptors and induction of intercellular adhesion molecule (ICAM-1) on cultured keratinocytes. J Cell Physiol. 1992;150(2):264–268. doi: 10.1002/jcp.1041500207. [DOI] [PubMed] [Google Scholar]
- 26.Kohrt HE, Colevas AD, Houot R, Weiskopf K, Goldstein MJ, Lund P, et al. Targeting CD137 enhances the efficacy of cetuximab. The Journal of clinical investigation. 2014;124(6):2668–2682. doi: 10.1172/JCI73014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lopez-Albaitero A, Lee SC, Morgan S, Grandis JR, Gooding WE, Ferrone S, et al. Role of polymorphic Fc gamma receptor IIIa and EGFR expression level in cetuximab mediated, NK cell dependent in vitro cytotoxicity of head and neck squamous cell carcinoma cells. Cancer immunology, immunotherapy : CII. 2009;58(11):1853–1864. doi: 10.1007/s00262-009-0697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3(12):952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 29.Johnson DR. Locus-specific constitutive and cytokine-induced HLA class I gene expression. J Immunol. 2003;170(4):1894–1902. doi: 10.4049/jimmunol.170.4.1894. [DOI] [PubMed] [Google Scholar]
- 30.Balsamo M, Vermi W, Parodi M, Pietra G, Manzini C, Queirolo P, et al. Melanoma cells become resistant to NK-cell-mediated killing when exposed to NK-cell numbers compatible with NK-cell infiltration in the tumor. Eur J Immunol. 42(7):1833–1842. doi: 10.1002/eji.201142179. [DOI] [PubMed] [Google Scholar]
- 31.Jamil KM, Khakoo SI. KIR/HLA interactions and pathogen immunity. J Biomed Biotechnol. 2011:298348. doi: 10.1155/2011/298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neisig A, Wubbolts R, Zang X, Melief C, Neefjes J. Allele-specific differences in the interaction of MHC class I molecules with transporters associated with antigen processing. J Immunol. 1996;156(9):3196–3206. [PubMed] [Google Scholar]
- 33.Neefjes JJ, Ploegh HL. Allele and locus-specific differences in cell surface expression and the association of HLA class I heavy chain with beta 2-microglobulin: differential effects of inhibition of glycosylation on class I subunit association. Eur J Immunol. 1988;18(5):801–810. doi: 10.1002/eji.1830180522. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Heron DE, Flickinger JC, Rwigema JC, Ferris RL, Kubicek GJ, et al. A retrospective, deformable registration analysis of the impact of PET-CT planning on patterns of failure in stereotactic body radiation therapy for recurrent head and neck cancer. Head & neck oncology. 2012;4:12. doi: 10.1186/1758-3284-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade Filho PA, Ito D, Deleo AB, Ferris RL. CD8+ T cell recognition of polymorphic wild-type sequence p53(65-73) peptides in squamous cell carcinoma of the head and neck. Cancer immunology, immunotherapy : CII. 59(10):1561–1868. doi: 10.1007/s00262-010-0886-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrade Filho PA, Lopez-Albaitero A, Gooding W, Ferris RL. Novel immunogenic HLA-A*0201-restricted epidermal growth factor receptor-specific T-cell epitope in head and neck cancer patients. J Immunother. 33(1):83–91. doi: 10.1097/CJI.0b013e3181b8f421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Albaitero A, Mailliard R, Hackman T, Andrade Filho PA, Wang X, Gooding W, et al. Maturation pathways of dendritic cells determine TAP1 and TAP2 levels and cross-presenting function. J Immunother. 2009;32(5):465–473. doi: 10.1097/CJI.0b013e3181a1c24e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveras-Ferraros C, Vazquez-Martin A, Queralt B, Adrados M, Ortiz R, Cufi S, et al. Interferon/STAT1 and neuregulin signaling pathways are exploratory biomarkers of cetuximab (Erbitux(R)) efficacy in KRAS wild-type squamous carcinomas: a pathway-based analysis of whole human-genome microarray data from cetuximab-adapted tumor cell-line models. International journal of oncology. 2011;39(6):1455–1479. doi: 10.3892/ijo.2011.1155. [DOI] [PubMed] [Google Scholar]
- 39.Garrido G, Rabasa A, Garrido C, Lopez A, Chao L, Garcia-Lora AM, et al. Preclinical modeling of EGFR-specific antibody resistance: oncogenic and immune-associated escape mechanisms. Oncogene. 2013 doi: 10.1038/onc.2013.288. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee-Kishore M, Kishore R, Hicklin DJ, Marincola FM, Ferrone S. Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J Biol Chem. 1998;273(26):16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19(15):4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer discovery. 2013;3(12):1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.