Figure 4. Proteomic Analysis of Interaction between PPARα and Simvastatin.
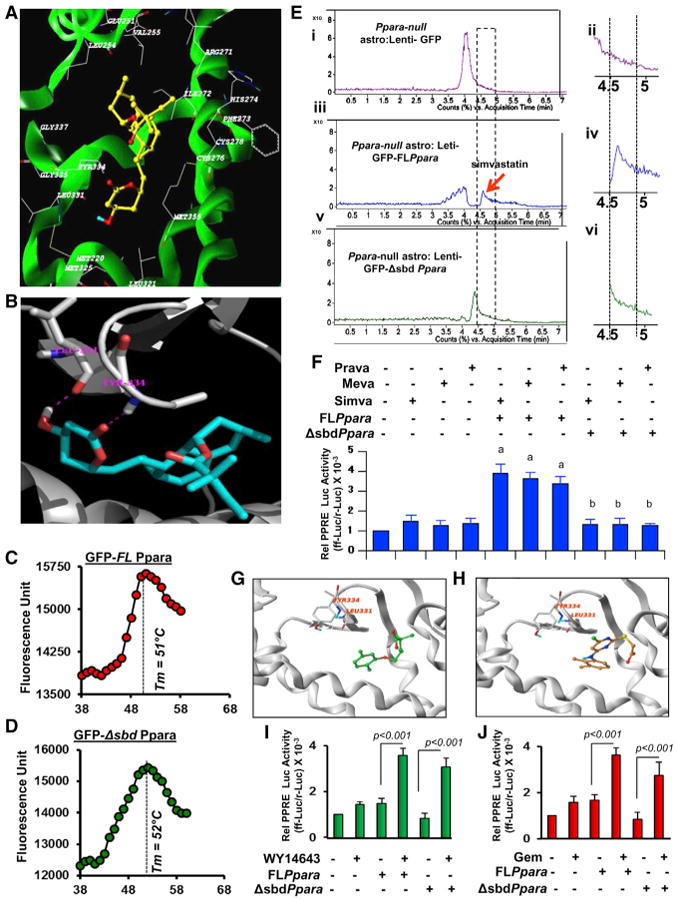
(A) Ribbon representations of the docked complex of PPARα (3VI8.pdb, X-ray, green) and simvastatin show the distribution of amino acids within a 5.00-Å distance of simvastatin.
(B) Magnified view of the docking site shows the interaction between simvastatin and Leu331 and Tyr334.
(C and D) Thermal shift assay of (C) full-length (GFP-FLPpara) and (D) statin-binding domain mutated (GFP-ΔsbdPpara) PPARα was performed.
(E) De novo binding assay of simvastatin where mouse astrocytes transduced with FLPpara and ΔsbdPpara constructs were treated with simvastatin for 2 hr, nuclear extracts were isolated, passed through GFP column for affinity-purification, and then analyzed by ESI-MS. Representative counts versus acquisition time ratio of simvastatin analyzed in the nuclear extracts of GFP-transduced (i and ii), GFP-FLPpara-transduced (iii and iv), and GFP-ΔsbdPpara-transduced (v and vi) astroglial cells.
(F) PPRE-driven luciferase activity in FLPpara and ΔsbdPpara-transduced Ppara null astrocytes after treatment with simvastatin (top), pravastatin (middle), and mevastatin (bottom). Results are mean ± SD of three independent experiments. ap <0.001 versus control; bp < 0.001 versus FLPpara.(G and H) Docked poses of gemfibrozil (G) and WY14673 (H) in PPARα ligand binding core.
(I and J) PPRE-driven luciferase activity in FLPpara and ΔsbdPpara-transduced Ppara null astrocytes after treatment with WY14643 (I) and gemfibrozil (J). Results are mean ± SD of three independent experiments.
