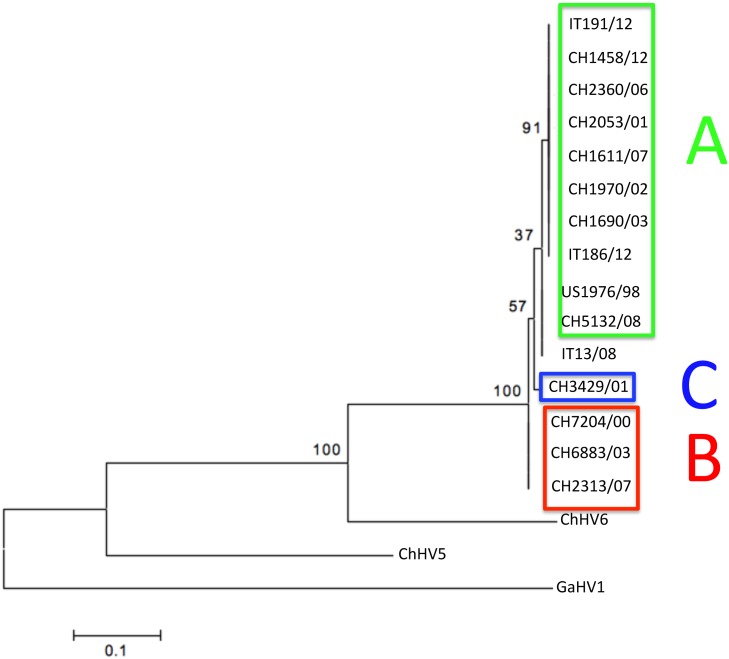
Fig 9. Glycoprotein B-based phylogenetic analysis.
Maximum likelihood tree inferred from the partial aa sequence of the gB of the 15 TeHVs strains investigated in this study and of reference strains of other ChHV (ChHV5, -6). Gallid herpesvirus 1 (GaHV1) was included as outgroup. Bootstrap values from 500 iterations are shown. This analysis reveals the existence of at least two distinct genogroups within the TeHV3 genotype (A-green boxed and B-red boxed). A third putative genogroup (C-blue boxed) composed of a single strain is also observed. A TeHV1 strain (IT13/08) clusters within the A genogroup of the TeHV3 genotype. Clear separation of the TeHV3 genogroups from the sequences of other two chelonian herpesviruses (ChHV5 and ChHV6) is observed. (Genbank accession numbers: CH1970/02-KP979727, CH1690/03-KP979726, CH1611/07-KP979721, CH2360/06-KP979725, CH2053/01-KP979723, CH5132/08-KP979729, CH7204/00-KP979719, CH6883/03-KP979730, CH2313/07-KP979722, CH3429/01-KP979720, CH1458/12-KP979718, US1976/98-KP979717, IT186/12-KP979728, IT191/12-KP979716, IT13/08-KP979724, ChHV5-AAU93326, ChHV6-AAM95776, GaHV1-YP_182356).