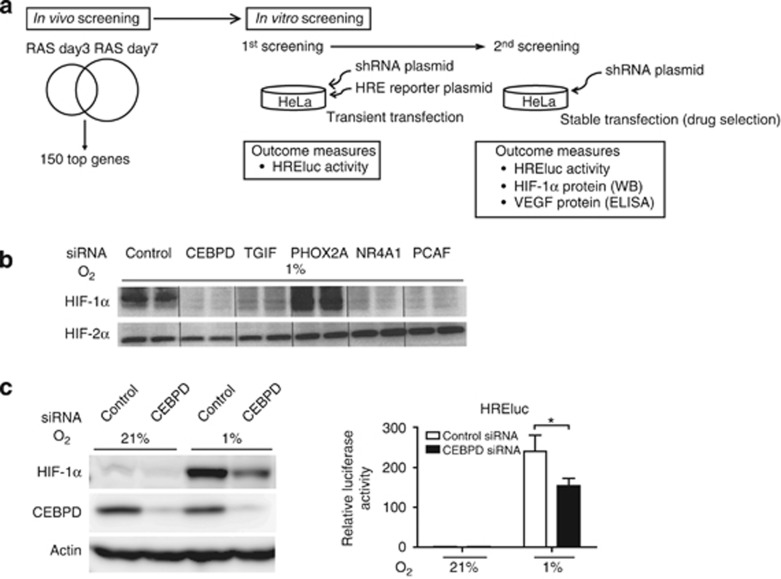
Figure 1.
The identification of hypoxia-inducible factor (HIF)-1-regulating genes. (a) Scheme of short hairpin RNA (shRNA) library screening. (b) HeLa cells transfected with small interfering RNA (siRNA) against the five candidate genes, CCAAT/enhancer-binding protein δ (CEBPD), transforming growth factor-beta-induced factor (TGIF), nuclear receptor superfamily 4A member 1 (NR4A1), paired mesoderm homeobox protein 2A (PHOX2A), or P300/CBP-associated factor (PCAF), were treated under hypoxia for 16 h to confirm the shRNA study results. The cells were processed for immunoblot analysis of HIF-1α and HIF-2α. Four genes except for PHOX2A showed a reduction in HIF-1α protein expression under hypoxia. The data are representative of three independent experiments, shown in duplicate. (c) Immunoblot analyses of HIF-1α (left panel) and HREluc activity (right panel) are shown representatively for siRNA-mediated CEBPD knockdown in HeLa cells. Knockdown of CEBPD decreased HIF-1α and hypoxic induction of HREluc activity in HeLa cells. (right panel) The ratio of luciferase reporter activity (firefly/TK-Renilla) to that of control siRNA under normoxia is indicated. Bar graph (mean±s.e.m. from three independent experiments) statistics performed using two-way analysis of variance (ANOVA) with Bonferroni post-hoc tests. *P<0.05. RAS, renal artery stenosis.