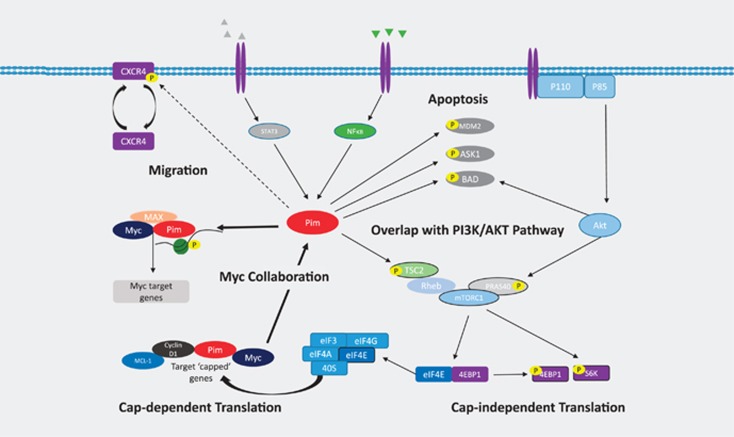
Figure 2.
Pim signaling in MM. Pim-2 is upregulated in MM. BMSCs, present in the microenvironment secrete IL-6 (depicted in grey) and increase Pim-2 transcription via STAT3 signaling. OCs release tumor necrosis factor family members TNFα, BAFF and APRIL (depicted in green), which then act via NFκB to increase Pim-2 transcription. Pim-2 has a role in prevention of apoptosis by phosphorylating MDM2 and reducing the degradation of p53, phosphorylation of ASK-1 and phosphorylation of the pro-apoptotic BAD at serine 112. This latter effect on BAD is shared with the PI3K/AKT/mTOR pathway also important in MM. These two pathways also converge on mTOR signaling. Pim-2 phosphorylates TSC2 to release the inhibitory effect of Rheb on mTORC1. Akt phosphorylates PRAS40 to activate mTORC1. Downstream of mTOR activation, both cap-dependent and cap-independent translation is initiated. The ribosomal proteins 4EBP1 and S6 kinase are phosphorylated to initiate cap-independent translation. Following phosphorylation of 4EBP1, eIF4E is released and forms a translation initiation complex with eIF4A, eIF4G and eIF3. The ribosomal 40S subunit can then bind to ‘weak' mRNA transcripts which contain a GC-rich region and capped by 7-methylguanosine. Among the pro-myeloma proteins translated in this manner are MYC, cyclin D1, MCL-1 and the Pim kinases themselves. This forms part of the oncogenic collaboration between the Pims and MYC. The Pims cannot perform oncogenic functions in the absence of MYC expression,39 and in turn, the Pims phosphorylate and stabilize MYC.40 Pims complex with MYC and MAX and are recruited to the E-box of MYC where Pims phosphorylate serine10 of histone 3 (H3S10) to induce transcription of up to 20% of MYC target genes.41 A putative role for the Pim kinases in MM, as has been demonstrated in other hematologic malignancies, is phosphorylation of CXCR4 on serine 339 with resultant internalization and re-expression of CXCR4, facilitating homing and migration.