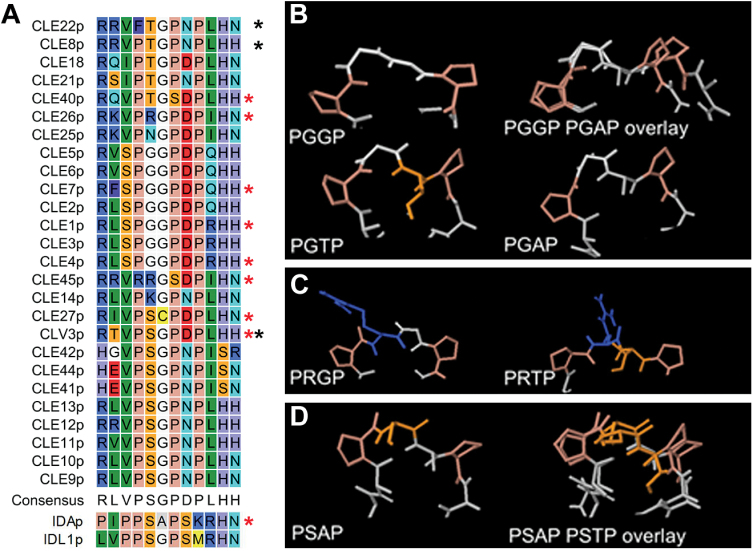
Fig. 6.
Peptide structure. (A) Manually adjusted alignment of the 12 amino acids of CLE peptides, with IDA and IDL1 for comparison. Red stars mark peptides used in the present study, black stars those tested by Song et al. (2013). (B–D) Examples of structures of the PXXP core predicted by PEP-FOLD. (B) PGGP. The larger Thr might interfere with receptor binding, and substitution of Gly6 with Ala6 may change the angles between the Pro residues. (C) PRGP. The side chain of Arg may change direction when the Gly6 is exchanged with a Thr. (D) PSAP. A change from Ala to Thr in position six may not result in major conformational changes when Ser is present in the core. (This figure is available in colour at JXB online.)