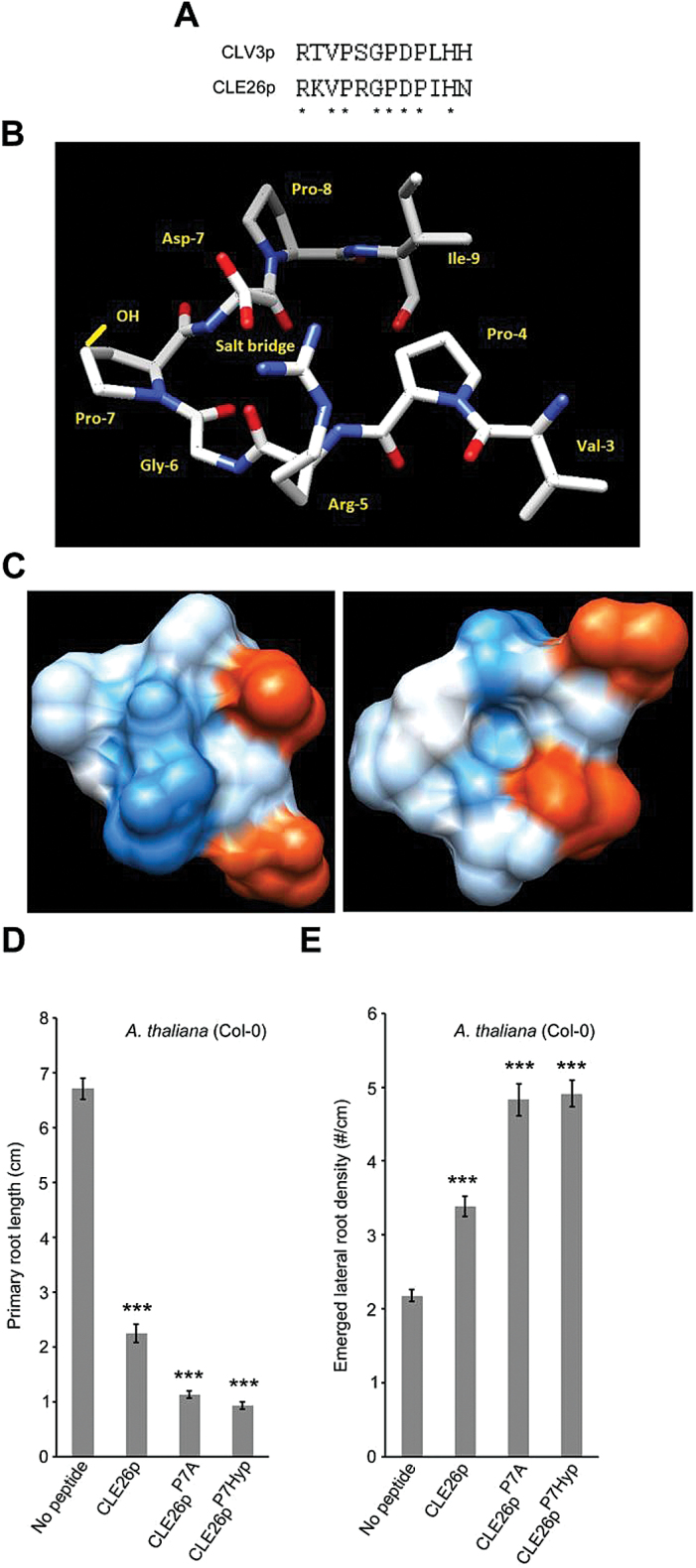
Fig. 5.
Sequence and structure versus activity of CLE26p. (A) Conserved residues between CLE26p and CLV3p are denoted by asterisks. (B) The top-ranked predicted structure with amino acids of the cleaved CLE26 peptide named, the position of a potentially stabilizing salt bridge marked, and the hydroxyl group of Pro-7-Hyp (in the 2S, 4S conformation reported from other studies) depicted in yellow. (C) The solvent-accessible surface (left) and solvent-accessible surface of the opposite face of the peptide in (B) (right) coloured in shades of red or blue to indicate the level of acidity or alkalinity, respectively. (D, E) Quantification of primary root length (D) and emerged lateral root density (E) for CLE26p, CLE27pP7A (~mCLE26p A7), and CLE26p7Hyp-treated wild-type seedlings. The graph indicates the mean ±SE. Statistical significance (Student’s t-test) compared with no peptide treatment is indicated: ***P<0.01.